Biology Group. 2024 June 28;5(6):653-659. doi: 10.37871/jbres1939.
Screening and Identification of Streptomyces lavendulae against Walnut Rot Disease Cytospora chrysosperma
Zhijin Liu, Lulu Shan, Shasha Xu, Rui Zhang and Xiaofei Chen*
- Walnut rot disease
- Antagonistics
- Identification
Abstract
Streptomyces is a biocontrol agent capable of protecting walnut from stem rot disease caused by the fungal pathogen Cytospora chrysosperma. Antagonistic strains were selected from walnut rhizosphere soil for biocontrol of the disease. The strains were screened using C. chrysosperma as the target pathogen. Using dilution coating plate separation method, plate confrontation method, growth rate method and in vitro branch protection experiment, screening the antagonistic actinobacteria with inhibitory effect and control effect on walnut tree rot disease. Strains were identified based on their morphological, physiological, and biochemical characteristics, 16S rDNA sequencing, and phylogenetic analysis. The strain F-02 was identified as Streptomyces Lavendulae. Plate confrontation method tests showed that the inhibition rates of the Streptomyces strain F-02 on the pathogens were approximately 80.30%. Further study showed that the filtrate of strain F-02 couldinhibit the C.chrysosperma, and the widths of their inhibition zone reached 1.24±0.15 mm, the prevention effect of 71.03%. For the first time, S.lavendulae was used for the biological control of walnut tree rot. It provides strain resources for the development of walnut rot agents. It has potential applications for biological control of walnut tree rot disease.
Introduction
Walnut (Juglans regia L.) is a significant economic tree species with in the walnut genus Pidaceae and ranks as one of the world's four major dried fruits, boasting rich nutritional and medicinal value [1]. Xinjiang stands as the second-largest province for walnut cultivation in China, with a planting area that reached 4.14x105hm2 and an annual output of 1.15x106t in 2020 [2]. However, in recent years, the extensive walnut planting in Xinjiang has been plagued by walnut rot disease, significantly impeding the healthy growth of the walnut industry in the region [3-5]. Presently, disease management relies heavily on chemical control methods, but these chemical fungicides pose various issues, including environmental pollution, risks to human and livestock safety, and pesticide residues [6,7]. Therefore, the development of efficient biological and fungal strains to replace chemical agents in controlling walnut rot disease can not only effectively manage disease occurrence and progression but also mitigate the problems associated with chemical control.
At present, many actinomycetes, mainly Streptomyces, have a strong inhibitory effect on pathogenic actinobacteria, such as Streptomyces castaneoglobosus [6], S.alboflavus, Streptomyces albidoflavus [7], etc. Cui Yunlong study found that the antibacterial substances released by golden S.aureus S-921 can dissolve the apple rot bacteria, smear the fermentation liquid on the diseased site, and the disease spots can be cured by Gupte T, et al. [8]. A Streptomyces UK10 isolated from soil with antagonistic effects against a variety of plant pathogens [9]. Jianc X, et al. [10] found that the siniscin produced by Streptomyces violet strain (S. lavendulae var. hainanensis) had good control effect on soft rot of Chinese cabbage and rice white leaf blight. At present, there are few studies on the biological control of walnut rot disease in China, and there is almost nothing in the control of raw actinomyces. In this study, actinomycetes isolated from walnut rhizosphere soil and screened out actinomycetes with Cytospora chrysosperma plate, and laid the foundation for further development and utilization of actinobacterial strains.
Test Materials and Methods
Soil materials
Experimental soil was collected from Walnut Forest Farm in Wensu County, Aksu Prefecture, in May 2023. The walnut variety in the orchard is "Wen185", 30 years old, with a spacing of 5 m × 7 m. In the process of sampling, select walnut trees with uniform growth, remove soil 0-5 cm 1 m away from the trunk of the walnut tree,researcher takes 5-20 cm of soil with soil drilling, then collects a total of 20 soil samples,puts the soil sample into a sterile bag, and a spacing of 5m ×7m.In the process of, researcher selects walnut trees with uniform growth,removes soil 0-5cm 1m away from the trunk of the walnut tree, take 5-20cm of soil with soil drilling, then collects a total of 20 soil samples , puts the soil sample into a sterile bag, and keep it in 4℃ refrigerator.
Pathogen material
Cytospora chrysosperma, Valsa mali, Alternaria alternate, and Fusarium oxysporum are provided by the key laboratory of the comprehensive pest control corps in southern Xinjiang.
Culture media
PDB (200 g potato, 20 g glucose, 1000 ml distilled water); PDA medium (200 g potato, 20 g agar, 1000 ml distilled water); High-1 medium (20 g soluble starch, 0.5 g sodium chloride, 1 g potassium nitrate, dipotassium hydrogen phosphate 0.5 g, magnesium sulfate, 0.01 g ferrous sulfate, 20 g agar, 1000 ml distilled water, pH7~7.5) 121℃ sterilized.
Test method
Isolation of the antagonistic strains: A soil sample weighing 10.0 g was placed into a conical flask along with 90 mL of sterile water and glass beads. The mixture was shaken on a shaker for 30 minutes to create a suspension of the soil sample. This suspension was then diluted to a 10 mL solution, followed by 10-2, 10-3, and 10-4 sample dilutions. Subsequently, 100 μL of each diluted solution was absorbed into PDA medium for strain isolation. The strains were isolated through dilution and coating for 3-7 days, followed by plate purification, culturing, numbering, and 4℃ storage.
Preliminary screening of the antagonistic strains: Using the plate confrontation culture method, a cross was made at an equal distance (approximately 2.5 cm) from the pathogen on the PDA plate inoculated with C. chrysosperma. The control consisted only of the pathogen, with each fungus treated in three replicates. The cultures were maintained at a constant temperature of 26℃ for 3d. Once the control mycelium was close to filling the plate, colony morphology was observed, and colony diameter was measured to calculate the inhibition rate of colony growth. The inhibition rate was calculated using the formula:
Inhibition rate (%) = [(colony diameter of the control group—colony diameter of the test group)/ colony diameter of the control group]×100%.
Compound screening of antagonists: Strains with initial screening inhibition rate greater than 60% were selected and rescreened by the four-point confrontation method.3d C. chrysosperma colonies were grown using a sterilized punch (5 mm diameter), fed to the center of the new PDA plate (90 mm diameter), and then connected to four identical strains with a needle 2.5 cm from the center of the plate, 26℃ constant temperature, with C. chrysosperma on PDA, each treatment was repeated three times. When the control was full, the diameter of the pathogen was measured by the cross cross method, and the inhibition rate was calculated. The inhibition rate was calculated using the formula:
Inhibition rate (%) = [(colony diameter of the control group—colony diameter of the test group)/ colony diameter of the control group]×100%.
Identification of the antagonistic strains:
- The Morphological Identification: Morphological characteristics and physiological and biochemical determination of antagonistic actinobacteria: Actinomycetes were cultured in a 30℃ constant temperature incubator for 3 d, and observed colony morphology, color, size, bulge shape, and edge characteristics. A series of physiological and biochemical parameters including siderophore, chitin, gelatin liquefaction, esterase, and pH were determined for the antagonistic strains. And refer to the Common Bacterial System Identification Manual [11].
- Molecular biology identification: Using the extracted strain DNA as a template using the bacterial universal primer 27F (5’-AGAGTTTGATCCTGGCTCAG-3’)/1492R (5’-GGTTACCTTGTTACGACTT-3’) [12]. The amplification reaction was carried out in a 50 µl system consisting of 2 µl DNA template, 2 μl each of upper and lower primers, 25 µl Taq PCR Master Mix, and 19 µl ddH2O. PCR amplification conditions included predenaturation at 95℃ for 5 min, denaturation at 95℃ for 35 s, annealing at 55℃ for 35 s, extension at 72℃ for 2 min, and extension at 72℃ for 10 min. The amplified products were analyzed by 1% agarose gel electrophoresis and viewed using an integrated gel imager. Qualified amplified products were then sent for sequencing to Shanghai Biological Engineering Co., Ltd. A phylogenetic tree based on 16S genes was constructed using MEGA 11 software to determine the taxonomic status of the antagonistic strains.
Determination of the strain antibacterial profile: In this study, the apple tree rot pathogen V. mali, and the walnut tree brown spot pathogen A. alternata, and F. oxysporum were selected as target bacteria. Antagonistic strains were positioned 2.5 cm away from the pathogenic fungus in a symmetrical manner, with PDA culture medium inoculated with pathogen cake alone serving as the control. Each treatment was replicated three times. The plates were then incubated at a constant temperature of 26°C. Following the growth of colonies in the control group, The inhibition rate was calculated using the formula:
Inhibition rate (%) = [(colony diameter of the control group—colony diameter of the test group)/ colony diameter of the control group]×100%.
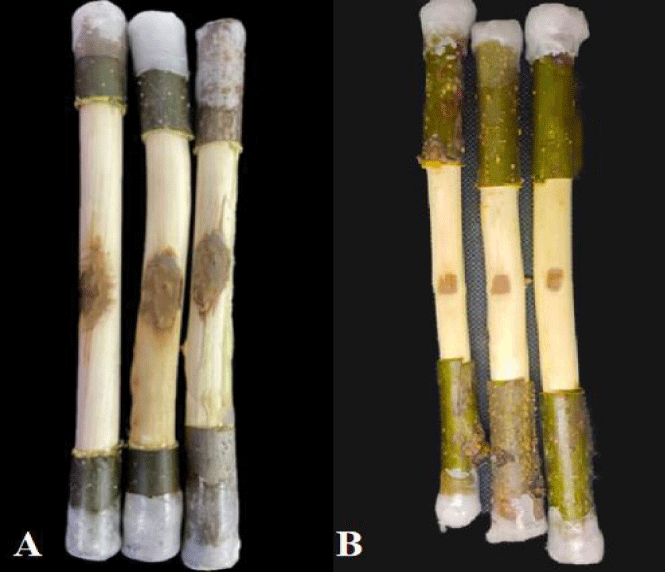
Prevention effect of biobacterial fermentation liquid on walnut rot in isolated branches: Punch holes in the branches with a sterilized 5 mm diameter hole punch, then inoculate each hole with a 5 mm diameter C. chrysosperma fungus cake (mycelial side facing down). Apply one inoculation point per branch. After incubating at a constant temperature of 26°C for two days, remove the pathogenic fungi cake using an inoculation needle. Dip a brush into an appropriate amount of antagonistic fungi fermentation liquid and apply it to the branches three times (allowing to air dry between each application). After moisturizing and cultivating at 26°C for 15 days, observe and measure the lesion areas. Use sterile water as a control, repeat the experiment three times, and calculate the control effect.
Disease spot area (cm2) =1 / 4 π long diameter and short diameter.
Prevention effect (%) = (control spot area-treated spot area) / control spot area ×100.
Data Analysis
The data of the trials were statistically and significantly analyzed by DPS software, and the phylogeny was constructed using with MEGA 11 software.3
Results and Analysis
Isolation and screening of the antagonistic strains
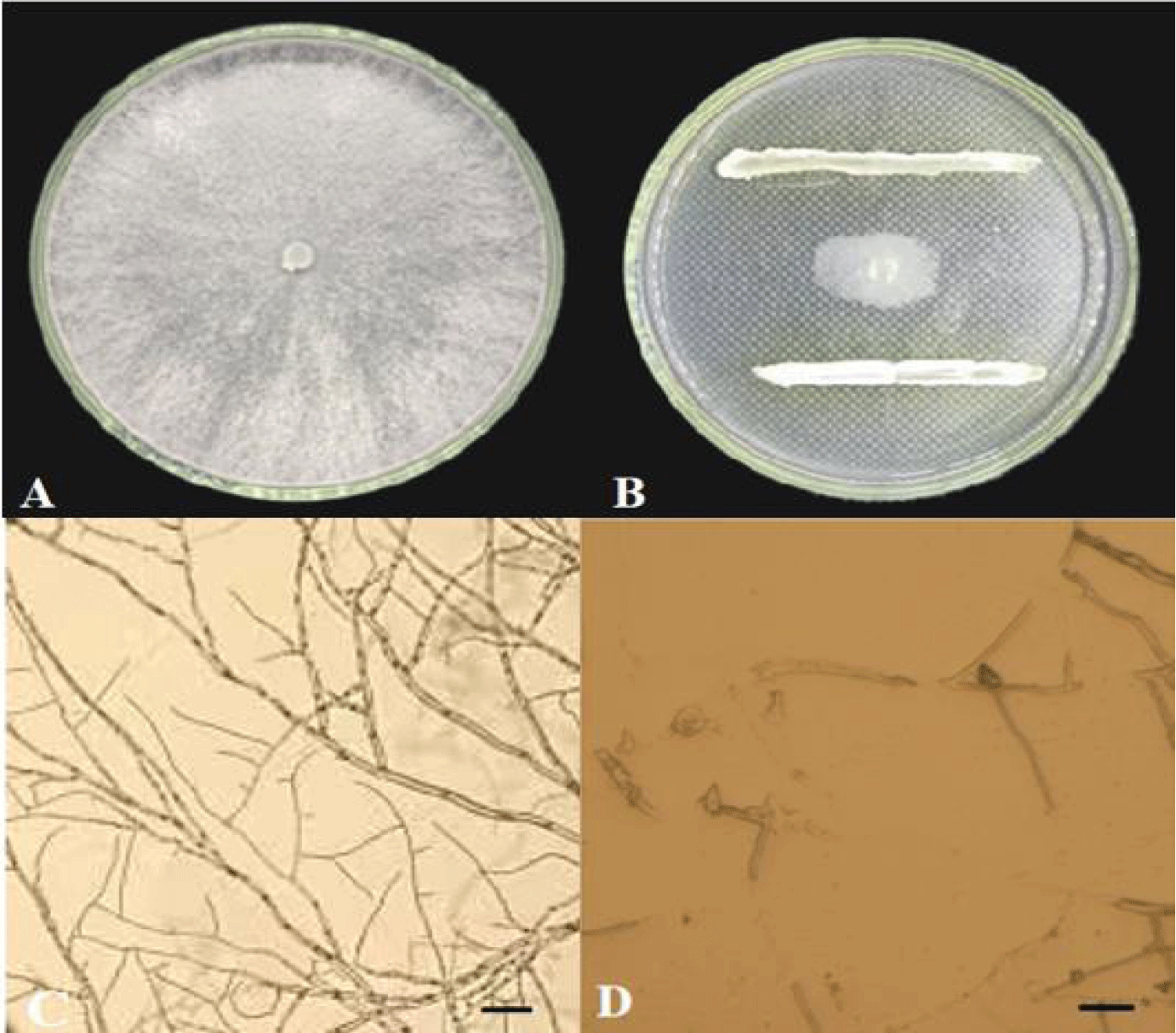
A total of 30 actinomycetes were isolated from walnut rhizosphere soil by dilution coated plate. Ten strains were antagonistic to C.chysosperma (Table 1). More than 60%, strain F-02 showed the strongest inhibition of C.chysosperma (Figure 1AB) with a inhibition rate of 80.30% (Table 2). By observing the effect of hyphal inhibition. After strain F-02 treatment, the pathogenic bacteria locally expanded and broke, and the color deepened and gradually blackened, and the growth of hyphae was inhibited (Figure 1D).The pathogenic bacteria of the control group were slender, with smooth surface, sharp top surface, complete structure and good growth (Figure 1C). Combined with primary and rescreening results, actinobacteria F-04 with high inhibition rate and stable antibacterial effect were selected for follow-up studies.
| Table 1: The screening results of the antagonistic strains. | |||||
| Strains | Colony diameter/cm | Inhibition rate / % | Strains | Colony diameter/cm | Inhibition rate / % |
| F-01 | 3.05 ± 0.15 | 61.88 ± 1.90 | F-06 | 3.31 ± 0.20 | 58.63 ± 2.34 |
| F-02 | 1.83 ± 0.10 | 77.13 ± 1.50 | F-07 | 3.60 ± 0.21 | 55.00 ± 1.88 |
| F-03 | 3.56 ± 0.11 | 55.50 ± 1.44 | F-08 | 3.36 ± 1.20 | 58.00 ± 3.60 |
| F-04 | 3.97 ± 0.20 | 50.38 ± 2.60 | F-09 | 2.31 ± 2.21 | 71.13 ± 4.60 |
| F-05 | 3.20 ± 0.15 | 59.96 ± 1.91 | F-10 | 3.54 ± 3.21 | 55.71 ± 5.60 |
| Note: The data listed in the table are average ± standard deviation. | |||||
| Table 2: Rescreening results of antagonistic strains against C.chysosperma. | ||
| Strains | Colony diameter/cm | Inhibition rate / % |
| F-01 | 3.29 ± 0.01a | 58.81 ± 0.08c |
| F-02 | 1.57 ± 0.04c | 80.30 ± 0.43a |
| F-09 | 2.35 ± 0.02b | 71.06 ± 0.27b |
| Note: The data listed in the table are mean ± standard deviation, and different lowercase letters represent whether the difference is significant in the <0.05 | ||
Identification of the antagonistic actinobacteria
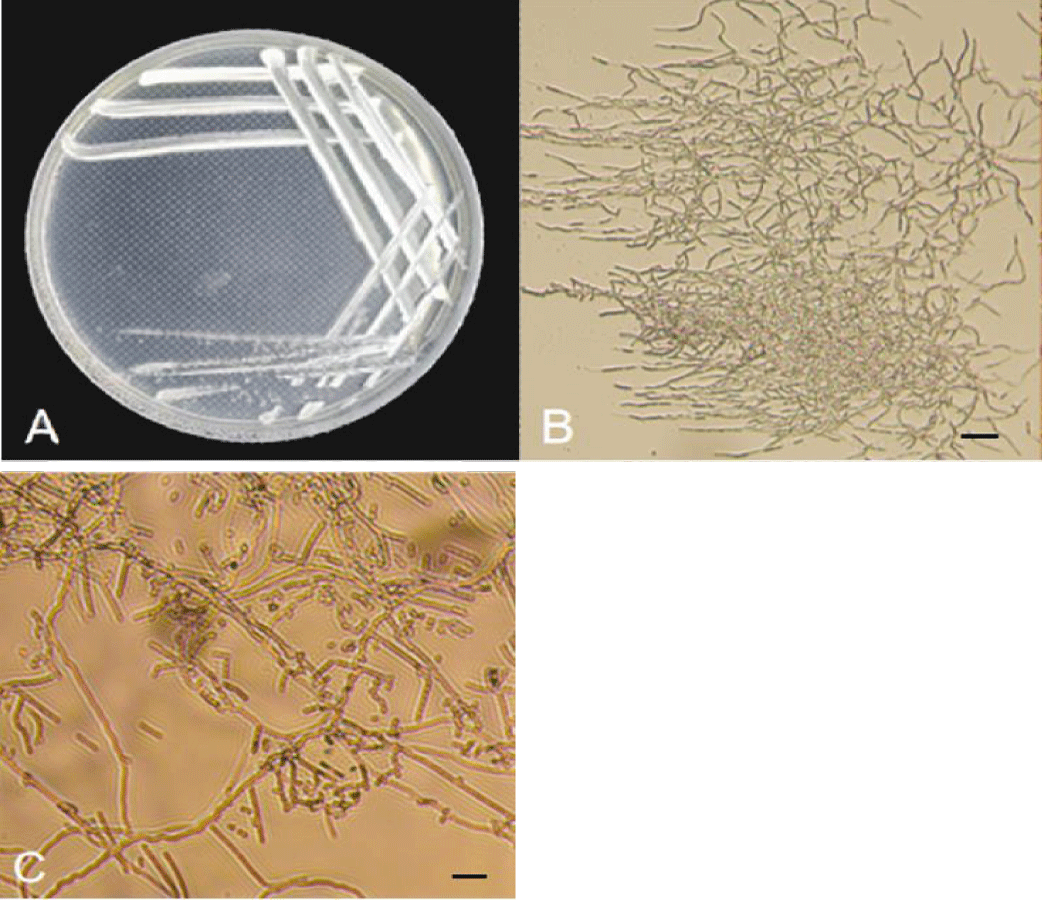
Mormorphological characteristics of antagonistic actinobacteria: Strain F-04 was single on High-1 solid medium, forming smaller, round, raised, light purple gray on the medium and pale yellow on the basal medium. The colonies grew slowly, the hyphae grew closely, and the spores were thin rods (Figure 2A-C). According to table 3, strain F-02 can use decomposed glucose, protein, fructose, histidine, KNO3, D-mannitol, (NH4) 2 SO4, methionine and leucine. The breakdown glutamate cannot be utilized (Table 3).
| Table 3: Physiological and biochemical determination of antagonistic bacteria. | |||
| Item | Result | Item | Result |
| Glucose | + | D-mannitol | + |
| Peptone | + | ( NH4 ) 2 SO4 | + |
| Fructose | + | Methionine | + |
| Histidine | + | Glutamate | - |
| KNO3 | + | Leucine | + |
| Note: "+" reaction is positive and ,"-" reaction is negative. | |||
Molecular and biological identification of antagonistic:
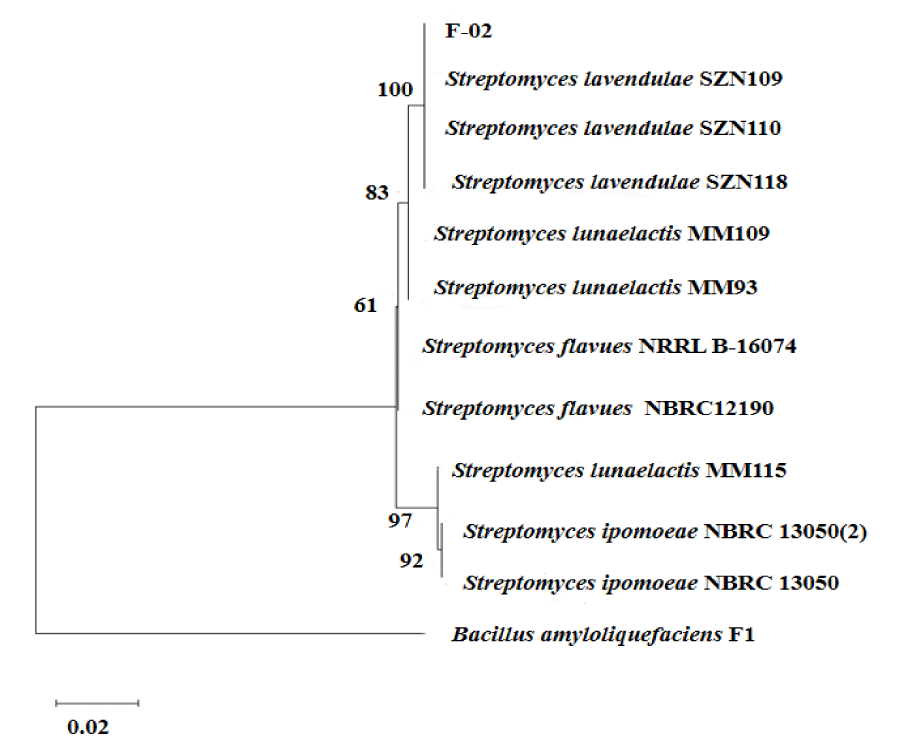
- Actinobacteria: Strain F-02 was sequenced by 16S, and sequence submission to GenBank obtained the accession number PP904437. Homology alignment analysis of amplified sequencing in the NCBI database showed that F-02 shares up to 100% identity with Streptomyces lavendulae. Using the MEGA11 software, a phylogenetic tree of 16S genes was constructed. As can be seen from figure 3, strain F-02 clustered in the same branch with S.lavendulae, which is the closest related to S.lavendulae. Based on morphological characteristics, physiological and biochemical characteristics, and molecular methods, strain F-02 was S.lavendulae.
Wide-spectrum determination of antagonistic actinobacteria
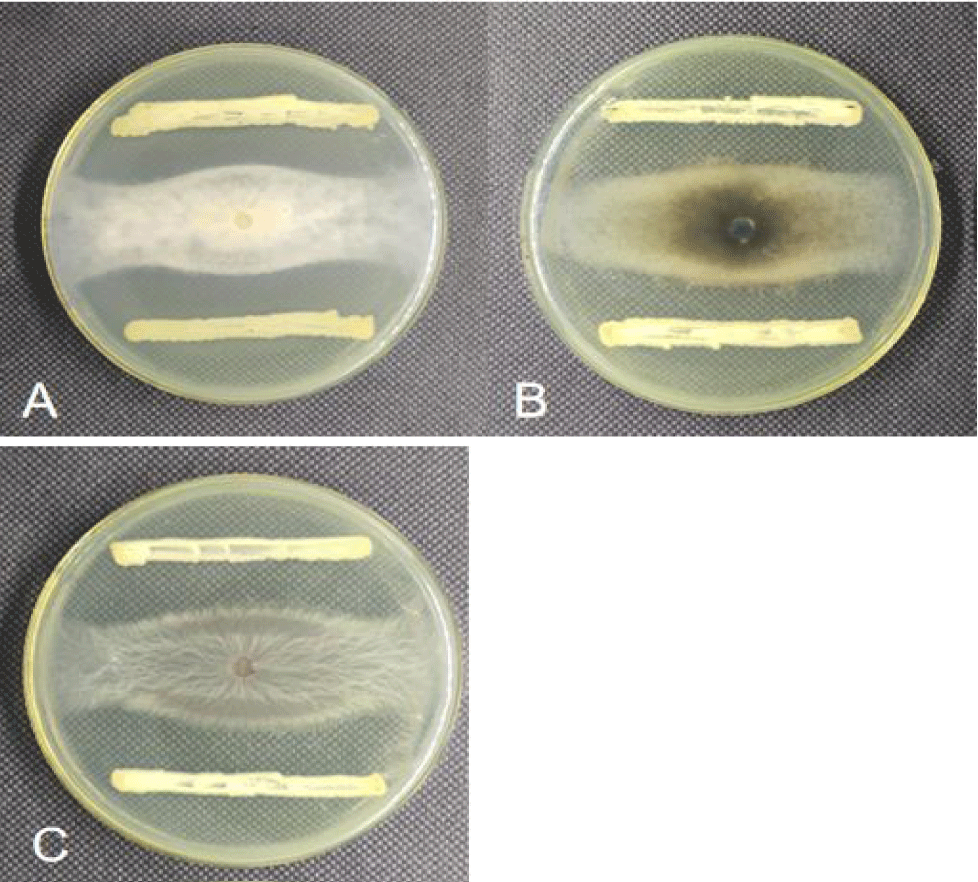
The inhibition effect of antagonistic strain F-02 on the three tested pathogens was determined by cross-plate confrontation. Figure 4 shows that the strains could effectively inhibit the growth of the three pathogens on the four sides. The calculation of the inhibition rate showed that strain F-02 inhibited more than 60% of the three pathogens (Table 4), with the best inhibition of V. mali, A. alternata and F. oxysporum: 73.13%, 67.50%, and 66.88%, respectively. Thus, it can be concluded that strain F-02 has good broad-spectrum resistance against plant pathogens.
| Table 4: Inhibition rate of antimicrobial bacteria on test pathogens. | ||
| Strains | Colony diameter /cm | Inhibition rate (%) |
| V. mali | 2.15 ± 0.15c | 73.13 ± 1.91a |
| A.alternata | 2.60 ± 0.10b | 67.50 ± 1.50b |
| F.oxysporum | 2.65 ±0.11a | 66.88 ± 1.44bc |
| Note: The data listed in the table are mean ± standard deviation, and different lowercase letters represent whether the difference is significant in the <0.05. | ||
The effect of biocontrol fermentation liquid on walnut rot spot
As can be seen from figure 5, strain F-02 had a good inhibitory effect on the expansion of walnut rot disease spot. The disease spot area of strain F-02 fermentation solution did not change significantly, and the disease spot area was 1.24cm2, while the control group reached 4.28cm2, which was significantly greater than the treatment group (p < 0.05). After calculation, the prevention effect of F-02 fermentation liquid on C.chysosperma was up to 71.03% (Table 5).
| Table 5: Protective effects of antagonistic bacterial fermentation broth on in vitro walnut shoots. | ||
| Strains | Lesion area (cm2) | Prevention effect (%) |
| CK | 4.28 ± 0.32a | - |
| F-02 | 1.24 ± 0.15b | 71.03 ± 0.15% |
| Note: The data listed in the table are mean ± standard deviation, and different lowercase letters represent whether the difference is significant in the <0.05. | ||
Discussion and Conclusion
Biological control is an effective means to realize the green prevention and control of plant diseases, and exploring the beneficial microorganisms with antagonistic effects to pathogens is the premise of biological control [13]. At present, there are studies on biological control of fruit rot in China. Wang Q, et al. [14] found that Streptomyces cavourensis has a good prevention and control effect on the apple tree decay disease caused by Valsa mali. There are few studies on the prevention of C. chrysosperma. In this study, a plate confrontation method was used to select a broad-spectrum antagonistic strain F-02. The antagonist strain F-02 was identified as Streptomyces lavendulae by morphological, physiological and biochemical characterization and molecular methods.
As an important microbial resource with biological research value, a large number of secondary metabolites of Streptomyces show good biological control effects on both bacterial and fungal diseases, which has become the main development direction of pesticides in the future. Liu Wei, et al. [15] found that the Streptomyces lavendulae gCLA4 strain has a good preventive effect on kiwi candis disease. Hu L, et al. [16] isolated and selected Sl-10 of rice from the rice rhizosphere. In this study, the use of Streptomyces lavendulae not only had a strong inhibitory effect on walnut rot actinobacteria, but also showed different degrees of inhibition on various pathogens, such as cotton wilt fungi and apple tree rot fungi, which were first reported in Xinjiang.
In this study, an antagonistic strain F-02 with broad-spectrum antibacterial effect was selected from walnut orchard soil in the Aksu region. Combining morphological characteristics, physiology, biochemistry, and l6S rRNA gene sequence identification, strain F-02 was identified as Streptomyces lavendulae.
References
- Qie Rongting, Zhang Yiping. Chinese fruit trees [M]. Beijing: China Forestry Press; 1996:59-63.
- The Xinxiang Uyghur autonomous region bureau of statistics. The Xinxiang statistical yearbook [M]. Beijing: China Statistics Press; 2022.
- Ma Rong, Wang Yangyang, Liu Xiaolin, Hu Jianxin, Tursunjiang Maimaiti Ali. Screening and preliminary identification of antagonistic bacteria in walnut tree rot disease in Xinjiang. Xinjiang Agricultural Science. 2015;52(05):895-901.
- Ma Yu, Ke Yang, Wang Qin, Li Bo, Li Yi. Symptoms of walnut ulcer disease and its pathogen identification [J]. Journal of Fruit Tree. 2014;31(3):443-447+525.
- Zhang Haijun, Chen Chunyan, Xie Yingping, Tian Zhongke. Isolation and identification of the pathogen of walnut rot disease [J]. China Plant Protection Guide. 2018;38(9):17-20.
- Li Zhitian, Cai Shulin, Wang He. ldentification of actinomycete strain qh-16 and structural analysis of lts active products[J]. Chinese Journal of Biological Control. 2021;37(01):156-164.
- Gao Zhenfeng, Zhao Jia. Study on antifungal properties of fermentation broth from streptomyces albidoflavus g-1 and optimization of lts fermentation condition. Biotechnology Bulletin. 2021;37(03):53-64.
- Cui Yunlong. Experimental study on the effect of S-921 strain and its fermentation broth on apple tree decay disease. Notification on biological control. 1992;8(1):26-28.
- Gupte T, Naik S. Isolation,taxonomic and fermentation studies on a new strain of Streptomyces cesarean var ukrainiana producing a tetraene antibiotic. World Journal of Microbiology and Biotechnology. 1999;15:545-552. doi: 10.1023/A:1008981528959.
- Jiang X L,Xie D L,Ni C F. The antibioticaction of zhongshengmycin. ActaPhytopathologica Sinica. 1997;27(2):40-45.
- Cai Miaoying, Dong Xiuzhu. Manual for systematic identification of common bacteria. Beijing: Science Press; 2001.
- Ma ZY, Wang JF, Zhu CW, Ping W. Isolation and characterization of filamentous bacteria HY10 accumu-lating poly-β-hydroxybutyrate (PHB) from activated sludge. Food and Fermentation Industries. 2012;38:101-105. doi: 10.13995/j.cnki.11-1802/ts.2012.05.001.
- MAO Tingting, Tao Gang, Zhao Xingli, Wang Qi, Li Shidong. Biological control of four kinds of microbial preparations against main diseases of pepper. Chinese Journal of Biological Control. 2020;36(2):258-264.
- Wang Qinghai, Wan Pingping, Li Anna. Inhibition of the metabolite of antifungal soil streptomyce spp.R15. China Agriculture Bulletin. 2006(2):327-330.
- Liu Wei, Wang Hua, Yang Ruolan. Effect of streptomyces lavendulae gcla4 on control of kiwifruit bacterial canker and ldentification of lts antibacterial substances. Journal of Northwest Agriculture, 2023;32(08):1289-1296.
- Hu Lingming, Xu Chunyi, Luo Jiaqi. Screening, identification and biocontrol effect of a strain of streptomyces lavendulae against Xanthomonas oryzae pv.oryzae. Journal of Plant Pathology. 2022;52(02):223-234.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.