Biology Group . 2023 April 04;4(4):590-599. doi: 10.37871/jbres1714.
Bioinformatic Analysis of the Association Between Myocardial Disease and Diesel Particulate Matter
Kyoung Jin Nho* and Jae Hoon Shin
- Diesel particulate matter
- Myocardial infarction
- H9C2 cells
- >Gene expression omnibus
Abstract
The etiological connection between Diesel Particulate Matter (DPM) and heart failure is unclear, despite the fact that multiple experimental studies have found a strong correlation between DPM and heart failure. We investigated the correlation between DPM and myocardial infarction using biological big data. Distinct gene expression profiles related to DPM and myocardial infarction were obtained from open-access genome databases, and a network analysis software was used to investigate their interaction. We investigated the detrimental effects of DPM on cardiac cells to understand the association between them. The putative signaling networks of 33 genes with differential expression were identified and studied. In addition to increasing the prediction accuracy, our study used experimental validation and literature-based analysis to enhance the knowledge of the mechanisms underlying DPM-related cardiac disease.
Abbreviations
DPM: Diesel Particulate Matter; GEO: Gene Expression Omnibus; DEGs: Differentially Expressed Genes; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; DAVID: Database for Annotation, Visualization, and Integrated Discovery; PPI: Protein-Protein Interaction
Introduction
Cardiac hypertrophy and ischemic injury (myocardial infarction) are major risk factors for heart failure and sudden death [1-3]. Pathophysiological stress in the heart causes physiological changes accompanied by transcriptional and epigenetic changes. This is characterized by increased cell size and extracellular matrix accumulation due to a variety of cardiovascular disorders, including pressure overload, heart valve dysfunction, and congestive heart failure [4-6]. Myocardial infarction continues to be one of the major causes of death worldwide despite significant improvements in therapy and prevention measures. Therefore, understanding the mechanisms underlying the development of cardiac hypertrophy, and myocardial infarction is essential for the prevention, and treatment of these diseases.
Diesel Particulate Matter (DPM) is a known carcinogenic factor and major air pollutant in cities [7,8]. The biological effect of particulate matter, such as DPM, is strongly related to its diameter. DPM majorly consisting of respirable particles less than 2.5 µm is classified as PM2.5. Exposure to DPM is a potential health hazard in large cities, where traffic is heavy resulting in air pollution and environments where diesel engines operate [9]. DPM is affected by fuel source, engine type, and operating conditions. Owing to its small particle size, DPM remains in the atmosphere for a long duration, adsorbing harmful substances such as various trace metals, Polycyclic Aromatic Hydrocarbons (PAHs), and nitroarenes, showing various chemical variabilities [10]. DPM enters cells, attaches to blood vessels, causes respiratory diseases, and move to other tissues through the blood, causing cardiovascular and lung function aggravation over long-term exposure, eventually leading to death [11]. Many studies have been reported on the effects of DPM on heart disease. According to previous studies, the correlation between increase in myocardial disease incidences and mortality, caused by exposure to DPM in the air has been clearly identified [12-14], but its biological pathogenesis remains unclear. Most previous studies have focused on the mechanism by which DPM exposure increases Reactive Oxygen Species (ROS) formation and the resulting inflammatory mediators expression [15,16]. However, studies on gene expression patterns are limited. Therefore, this study screened DPM and myocardial infarction data expression profiles from the Gene Expression Omnibus (GEO) database. Based on the analysis results, we selected genes involved in the myocardial disease network according to DPM treatment. Furthermore, verification experiments such as quantitative real-time-polymerase chain reaction (qRT-PCR) should be conducted and the genes involved in the induction and exacerbation of myocardial disease by DPM need to be identified.
Materials and Methods
Materials
Labware including multi-well plates, serological pipettes, and spreaders, were bought from BD Falcon (Franklin Lakes, NJ, USA) and Corning Incorporated (Corning, NY, USA). Paraformaldehyde (4%) and Triton X-100 (0.1%) were bought from Sigma-Aldrich (St. Louis, MO, USA).
Preparation of DPM
Standard Reference Material 1650b (SRM1650b) was purchased from the National Institute of Standards and Technology (NIST). Refer to NIST (1650b.pdf (nist.gov)) for SRM1650b certificate of analysis. SRM1650b was suspended in Phosphate-Buffered Saline (PBS) at 10 mg/mL stock concentration.
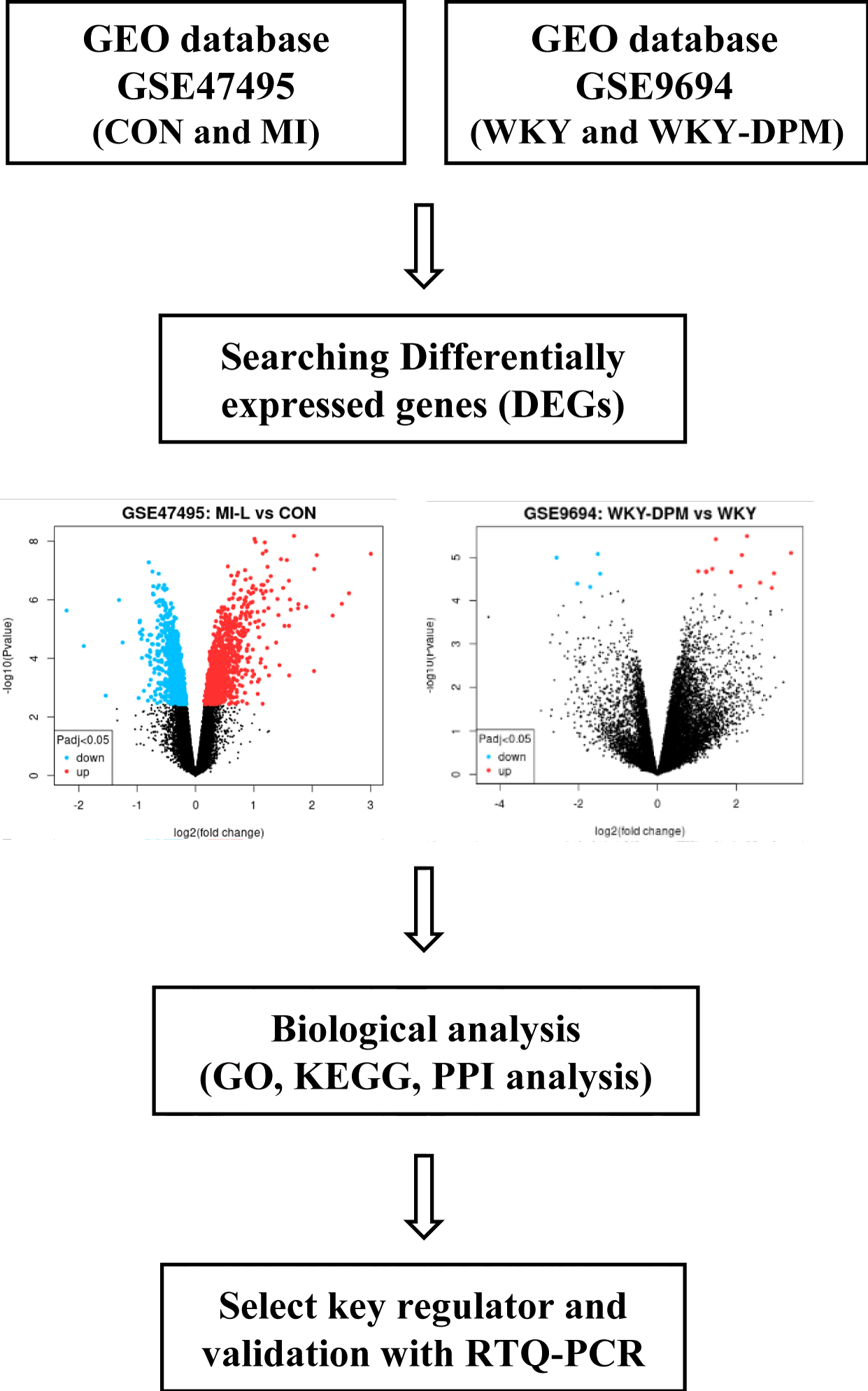
Searching data and identification of differentially expressed genes
Expression profiles of DPM and myocardial infarction were acquired from the Gene Expression Omnibus (GEO) database; two independent data series, GSE47495 and GSE9694, were used in our study. The following keywords were used to screen gene expression datasets: diesel particulate matter, gene expression profiles by array, Rattus norvegicus, cardiac hypertrophy, myocardial infarction, and heart failure. The details of the two data series are listed in table 1, and figure 1 displays the study design plan. GEO2R is an online tool that allows comparisons between two groups of samples to analyze the GEO database using the limma R package. We analyzed the DEGs as differentially expressed genes in GSE47495 under the condition of adj. p < 0.05 and log(FC) ≥ 1.5. In addition, GSE9694 was analyzed with an adj. p - value < 0.05, and log(FC) > 1.
| Table 1: Details for GSE47495 and GSE9694 data series. | ||||
| Accession No. | Platform | Sample | Organism | Group |
| GSE47495 | GPL6247 | Left ventricular tissue | Rattus norvegicus | CON = sham-operated rat (n = 6) MI = myocardial infarction (n = 5) |
| GSE9694 | GPL341 | Heart | Rattus norvegicus | WKY = wistar kyoto rat air exposure (n = 4) WKY-DPM = wistar kyoto rat diesel exposure (n = 4) |
Functional enrichment analysis
The molecular correlation between myocardial infarction and DPM was examined using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) and ShinyGO 0.77 online tools. A Shiny application called ShinyGO is built on a number of R/Bioconductor programs, as well as a sizable annotation and pathway database compiled from multiple sources. The functional categories of Biological Process (BP), Cell Components (CC), and Molecular Functions (MF) were included in the GO enrichment. By measuring and rating the gene count simultaneously, meaningful GO biological processes and pathway enrichment analyses were selected, with statistical significance set at p < 0.05. Data analysis and visualization were done using the ShinyGo enrichment tool.
PPI network
The online database Search Tool for the Retrieval of Interacting Genes/Proteins was used to analyze the Protein-Protein Interaction (PPI) network between myocardial infarction and DPM (STRING: functional protein association networks (string-db.org)).
Cell culture and treatments
H9C2 cells were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA) and propagated according to the instructions provided. Dulbecco′s Modified Eagle′s medium (DMEM) was used to sustain cells on a regular basis. It contains 10% fetal bovine serum from ATCC and 1% penicillin-streptomycin (Gibco BRL, Gaithersburg, MD, USA).
Cells were incubated at 37°C and 5% carbon dioxide. Before treatment, the stock solution was sonicated for 1h in an ultrasonic bath to adequately disperse particulate matter. The stock solution was diluted with DMEM for treatment. After treatment, each well was washed twice with PBS to reduce the effect of the remaining particles.
Immunofluorescence staining
Cell surface measurements followed previous studies [17]. Briefly, H9C2 cells cultured on the coverglass were washed with PBS and then fixed with 4% paraformaldehyde for 10 min. Again, washed twice with PBS, and permeabilized with 0.1% Triton X-100 in PBS. After blocking with 1% BSA in PBS for 1h at room temperature, cells were incubated with an anti-α-actinin antibody, conjugated to Alexa Fluor 488. 4',6-diamidino-2-phenylindole (DAPI) was used to stain the nuclei (Molecular Probes, Eugene, OR, USA). After washing, the slides were mounted using a fluorescent mounting medium. Axio Imager M2 microscope from Carl Zeiss (Oberkochen, Germany), was used to examine the prepared cells, and ZEISS ZEN 3.7 software was used for analysis (Carl Zeiss).
RNA extraction and quantitative real-time PCR (qRT-PCR)
H9C2 cells treated with DPM were prepared, and total RNA was extracted using PureLink RNA mini kit (Invitrogen, USA). For cDNA synthesis, High-Capacity RNA-to-cDNA Kit was used (Applied Biosystems, Waltham, MA, USA). The primer used for qRT-PCR shown in Table 2. QuantStudio 3 Real-Time PCR System and SYBR green dye from Applied Biosystems to analyze and quantify the expression levels of mRNA. The mRNA expression was normalized to 18S rRNA, and the comparative Ct (ΔΔCt) method was used for the expression level. All reactions were performed in triplicates.
| Table 2: Primer lists for qRT-PCR analysis. | |||
| Genes | Forward | Reverse | Accession number |
| Sfrp1 | CTGCCACCAGCTGGACAAC | ACCTTGCGCCCCATGAT | NM_001276712.2 |
| Sfrp2 | TGTCCGATAGGGACCTGAAGA | GGTGCACTGCAGGCTGTCT | NM_001100700.1 |
| Fzd2 | GGATCTGGTCCGGCAAGAC | GTTGGTGAGACGCGTGTAGAAC | NM_172035.2 |
| Atp2a2 | TTGGGTTTCCTGAAGCTTTGA | ACCATCCGTCACCAGATTGAC | NM_001110139.2 |
| Atp1a3 | CTGTCCTCGGGTTCCGTAAA | GGGAATCTCGGCCACTTTCT | NM_012506.1 |
| Nppa | ACCTGCTAGACCACCTAGAGG | GCTGTTATCTTCCGTACCGG | NM_012612.2 |
| Nppb | CAGCTGCCTGGCCCATCACT | ACCTCCCAGCGGCGACAGAT | NM_031545.1 |
| 18S | AAGTTTCAGCACATCCTGCGAGTA | TTGGTGAGGTCAATGTCTGCTTTC | NM_213557.1 |
Statistical analysis
All experimental results were expressed using mean ± standard error of the mean (SEM). For statistical analysis, an unpaired Student’s t test or one-way ANOVA method was used. Triplicates of each experiment were conducted. Compared to the control group, P < 0.05 was determined to be statistically significant.
Results and Discussion
Identification of DEGs
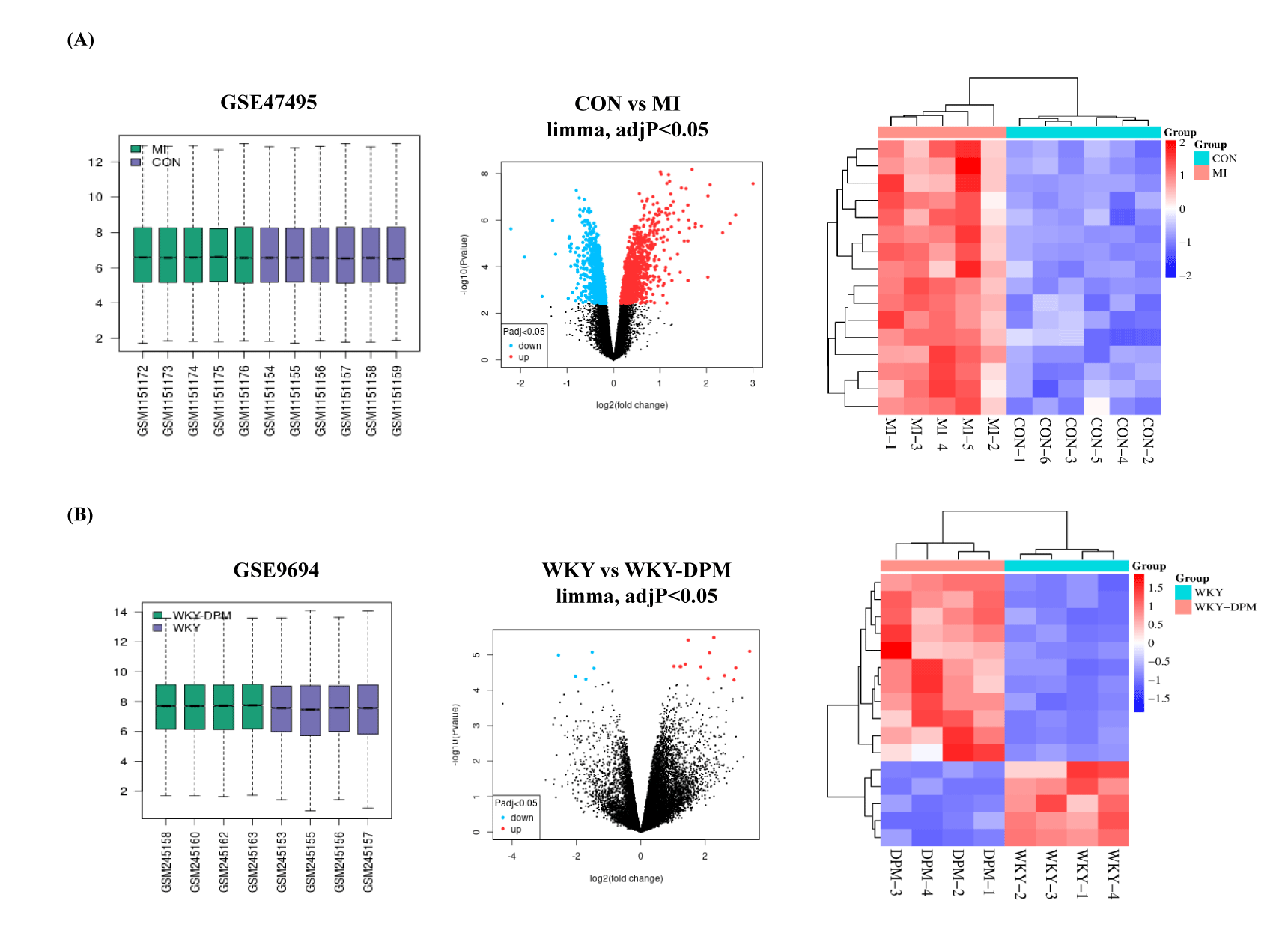
Microarray results of the GSE47495 and GSE9694 DEGs were screened using GEO2R. The details of the two data series are listed in table 1. GSE47495 included five left ventricular tissue samples from rats undergoing Myocardial Infarction (MI) surgery and six left ventricular tissue samples from rats undergoing a sham operation (CON). First, 1,649 DEGs were obtained from the GSE47495 data series (adj. p < 0.05, CON vs. MI), including 844 up-regulated and 805 down-regulated genes. Then, the top 18 up-regulated DEGs from the GSE47495 data series were acquired with the criteria of log(FC) ≥ 1.5 (Figure 2A and table 3). In addition, the GSE9694 data series obtained included four Wistar Kyoto (WKY) rats exposed to air or the whole body to diesel emission particles for 4 weeks (WKY-DPM). Fifteen DEGs were acquired from the GSE9694 dataset (WKY vs. WKY-DPM), including 11 up-regulated (log(FC) > 1) and four down-regulated (log(FC) < -1.5) with an adj. p < 0.05 (Figure 2B and Table 4). Then, we conducted a hierarchical clustering analysis to show DEGs as a heatmap using a SRplot (Figure 2).
| Table 3: Top 18 up-regulated DEGs form analysis of the GSE47495 data series. | |||||
| ID | adj.p-Val | p-Value | logFC | Gene symbol | Gene title |
| 10815369 | 0.000111 | 2.73E-08 | 3 | Postn | periostin |
| 10719728 | 0.000533 | 1.39E-06 | 2.5 | Atp1a3 | ATPase Na+/K+ transporting subunit alpha 3 |
| 10750505 | 0.000802 | 3.49E-06 | 2.35 | Pcp4 | Purkinje cell protein 4 |
| 10706059 | 0.000111 | 3.04E-08 | 2.07 | Sbsn | suprabasin |
| 10720829 | 0.00019 | 9.10E-08 | 2.03 | Hamp | hepcidin antimicrobial peptide |
| 10940654 | 0.009324 | 2.76E-04 | 2.03 | Spp1 | secreted phosphoprotein 1 |
| 10873899 | 0.000594 | 1.79E-06 | 1.89 | Nppa | natriuretic peptide A |
| 10792344 | 0.000533 | 1.42E-06 | 1.76 | Sfrp1 | secreted frizzled-related protein 1 |
| 10816144 | 0.000605 | 1.99E-06 | 1.75 | Sfrp2 | secreted frizzled-related protein 2 |
| 10764069 | 8.13E-05 | 6.70E-09 | 1.69 | Chi3l1 | chitinase 3 like 1 |
| 10717233 | 0.000644 | 2.20E-06 | 1.63 | Ctgf | connective tissue growth factor |
| 10842043 | 0.000466 | 9.89E-07 | 1.61 | Wisp2 | WNT1 inducible signaling pathway protein 2 |
| 10927361 | 0.000262 | 2.27E-07 | 1.6 | Ankrd23 | ankyrin repeat domain 23 |
| 10935041 | 0.001186 | 7.82E-06 | 1.6 | Tceal7 | transcription elongation factor A like 7 |
| 10917183 | 0.011489 | 3.88E-04 | 1.6 | Ncam1 | neural cell adhesion molecule 1 |
| 10928761 | 0.000131 | 4.48E-08 | 1.56 | Fn1 | fibronectin 1 |
| 10918869 | 0.001186 | 7.88E-06 | 1.52 | Col12a1 | collagen type XII alpha 1 chain |
| 10917883 | 0.000321 | 4.01E-07 | 1.5 | Loxl1 | lysyl oxidase-like 1 |
| adj.p-value < 0.05, log(FC) ≥ 1.5. | |||||
| Table 4: Top 15 DEGs from analysis of the GSE9694 data series. | |||||
| ID | adj.p-Val | p-Value | logFC | Gene.symbol | Gene.title |
| 1368609_at | 0.0294 | 0.00002337 | 2.96 | Slc10a1 | solute carrier family 10 member 1 |
| 1389706_at | 0.0453 | 0.00005118 | 2.90 | Dclk1 | doublecortin-like kinase 1 |
| 1373931_at | 0.027 | 0.00000326 | 2.28 | Pianp | PILR alpha associated neural protein |
| 1369091_at | 0.027 | 0.00000887 | 2.14 | Chrnb1 | cholinergic receptor nicotinic beta 1 subunit |
| 1386514_at | 0.0453 | 0.00004622 | 2.10 | Hapln4 | hyaluronan and proteoglycan link protein 4 |
| 1375539_at | 0.0294 | 0.00002184 | 1.87 | Tbr1 | T-box, brain, 1 |
| 1388093_at | 0.027 | 0.00000382 | 1.48 | Tas2r126 | taste receptor, type 2, member 126 |
| 1374587_at | 0.0294 | 0.0000185 | 1.39 | Tmtc3 | transmembrane and tetratricopeptide repeat containing 3 |
| 1374864_at | 0.0294 | 0.00002153 | 1.25 | Spry2 | sprouty RTK signaling antagonist 2 |
| 1375164_at | 0.0294 | 0.00002133 | 1.23 | Atp2a2 | ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| 1390432_at | 0.0294 | 0.00002109 | 1.03 | Tmco3 | transmembrane and coiled-coil domains 3 |
| 1372989_at | 0.027 | 0.00000843 | -1.51 | Zdhhc14 | zinc finger, DHHC-type containing 14 |
| 1398304_at | 0.0453 | 0.00004841 | -1.71 | Fzd2 | frizzled class receptor 2 |
| 1368368_a_at | 0.043 | 0.0000405 | -2.04 | Lsr | lipolysis stimulated lipoprotein receptor |
| 1390176_at | 0.027 | 0.00001019 | -2.56 | Fam84b | family with sequence similarity 84, member B |
| adj.p-value < 0.05, log(FC) >1 or < -1.5 | |||||
Function enrichment analysis and PPI network
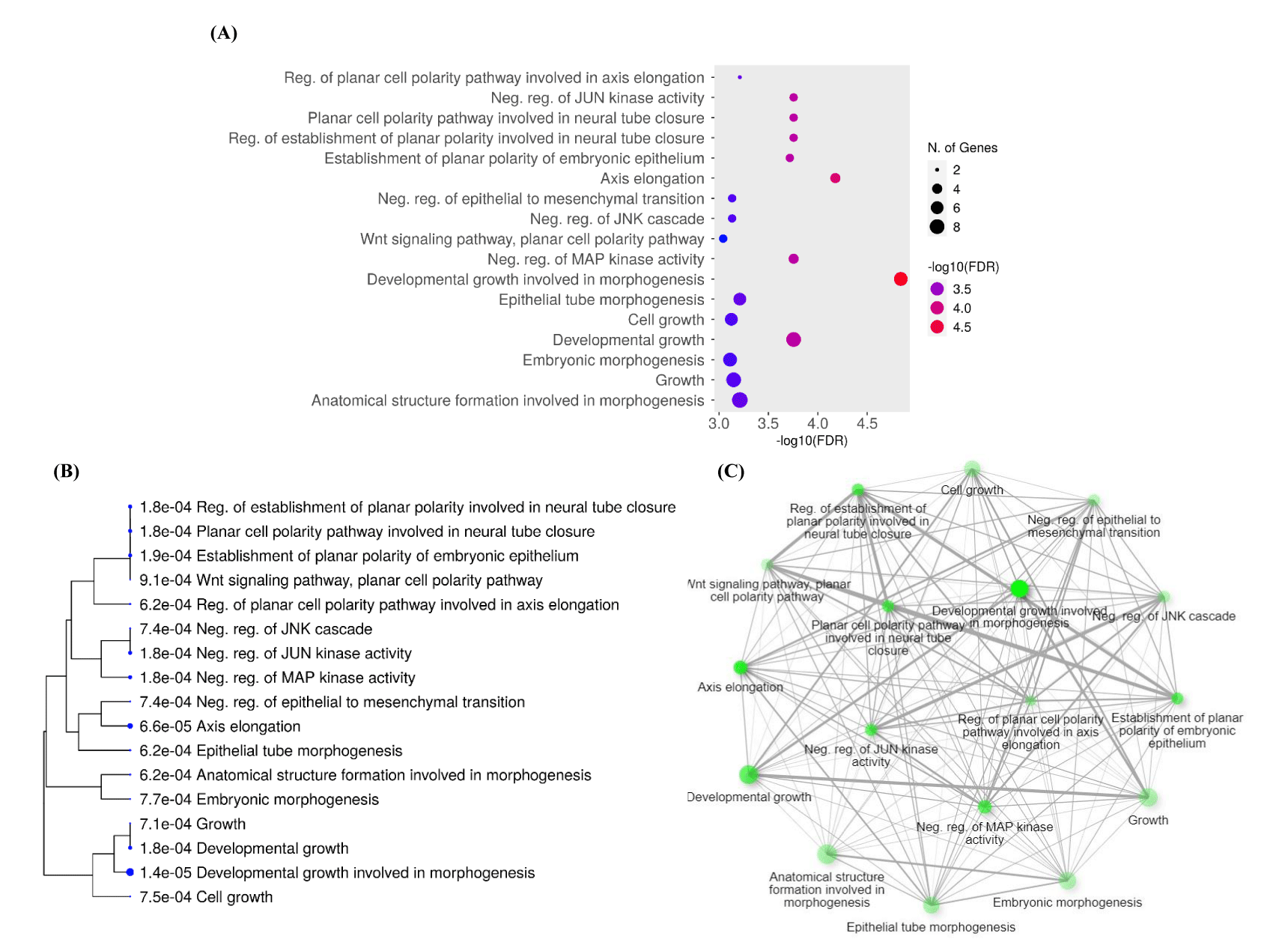
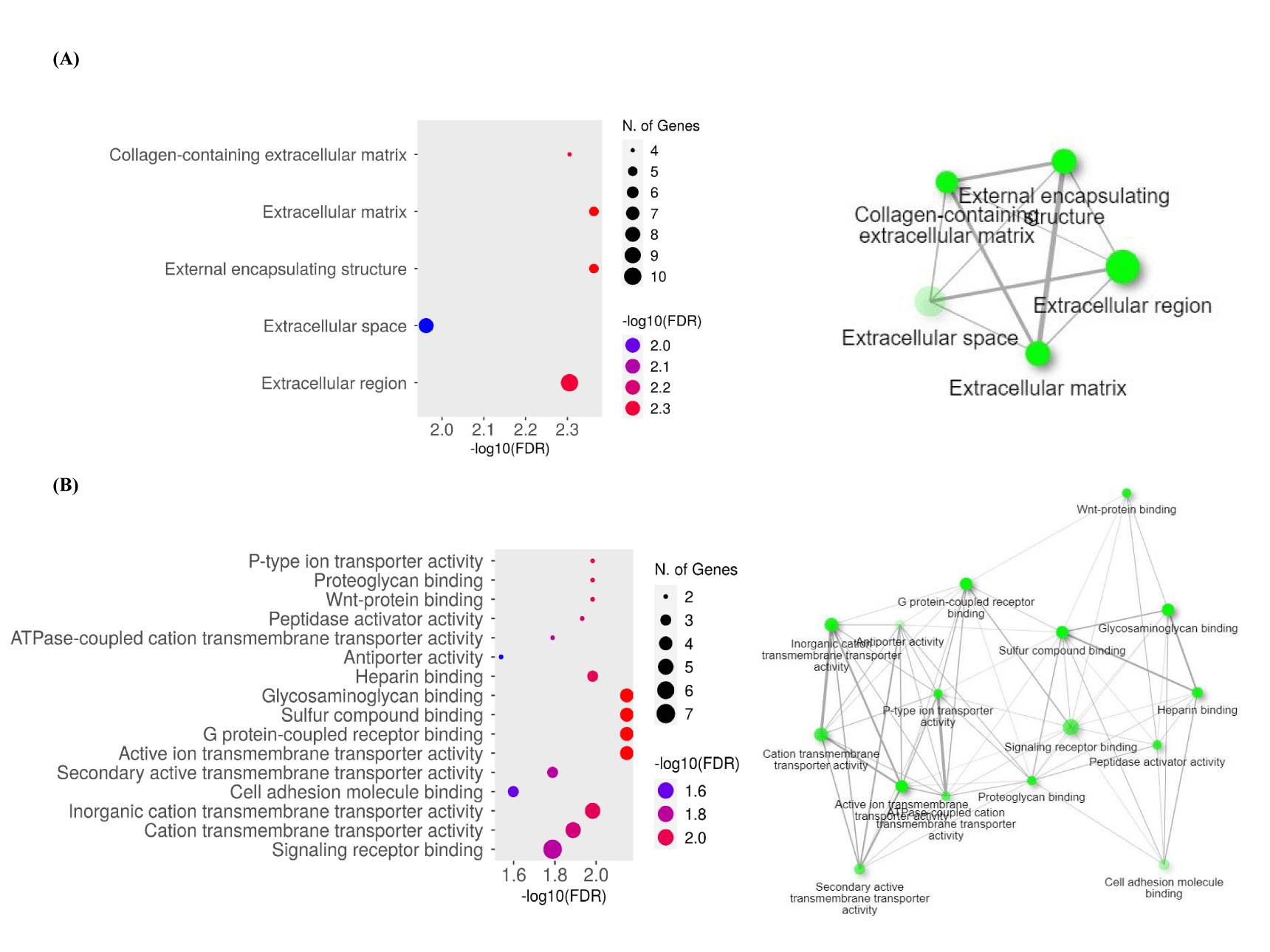
To determine the function of the target genes, we conducted GO and KEGG analyses of the interacting 33 DEGs from GSE47495 and GSE9694 using DAVID and ShinyGO 0.77 online tools. Three functional categories: Biological Process (BP), Cell Components (CC), and Molecular Functions (MF) were created from the results of the GO enrichment study. Following DEG analysis, significantly enhanced BP is displayed in a dot plot, clustering tree, and graphical network (Figure 3). The BP results indicated that DEGs were mainly enriched in developmental growth involved in morphogenesis, axis elongation, developmental growth, and negative regulation of Mitogen-Activated Protein Kinase (MAPK) activity (Figures 3A&B). BP with a large number of shared genes is grouped. The enriched terms were arranged into a network with edges connecting overlapping gene sets (Figure 3C). Furthermore, the extracellular region, external encapsulating structure, collagen-containing extracellular matrix, and extracellular matrix were enriched in the CC category (Figure 4A). The most enriched MFs were G protein-coupled receptor binding, glycosaminoglycan binding, inorganic cation transmembrane transporter activity, active ion transmembrane transporter activity, and sulfur compound binding (Figure 4B).
PPI network and KEGG analysis
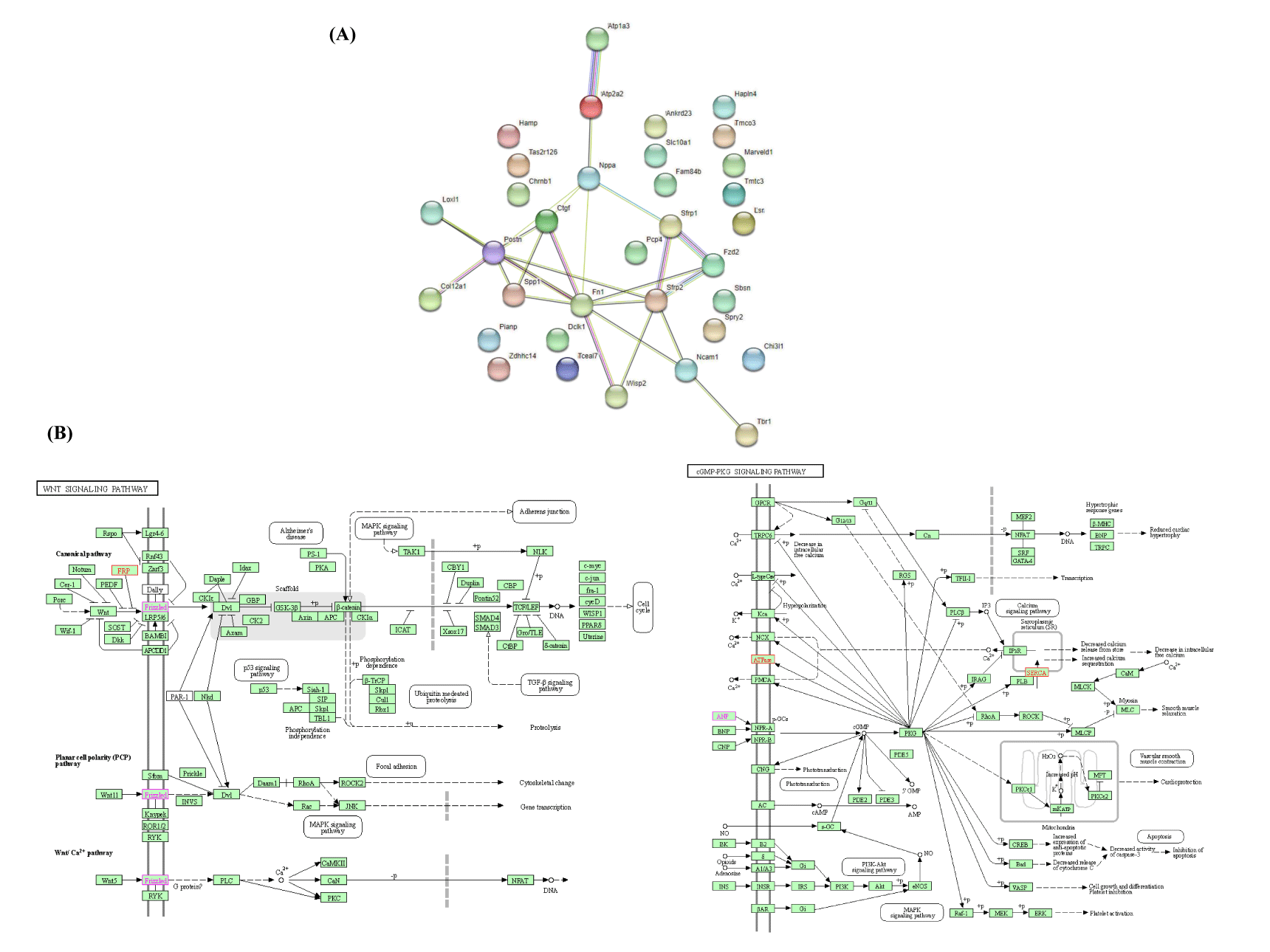
As shown in figure 5A, we mapped the PPIs using STRING to distinguish connections among the 33 genes. The network is composed of 33 nodes and 26 edges, and has an average local clustering coefficient of 0.285. The STRING database was used to analyze the numerous proteins, and the results showed that two biological signaling pathways, the Wnt and CGMP-PKG signaling pathways, were considerably impacted. Using DAVID, KEGG analysis simultaneously showed that the DEGs in CGMP-PKG and Wnt signaling pathways were highly enriched (Figure 5B). The gene list shown in table 5.
| Table 5: Significantly impacted signaling pathways in GSE47495 and GSE9694 data series. | ||||
| Enrichment FDR | Pathway Genes | Fold Enrichment | Pathway | Genes |
| 0.044 | 1583 | 14.08 | Wnt signaling pathway | Sfrp2, Sfrp1, Fzd2 |
| 0.044 | 164 | 13.56 | CGMP-PKG signaling pathway | Atp2a2, Atp1a3, Nppa |
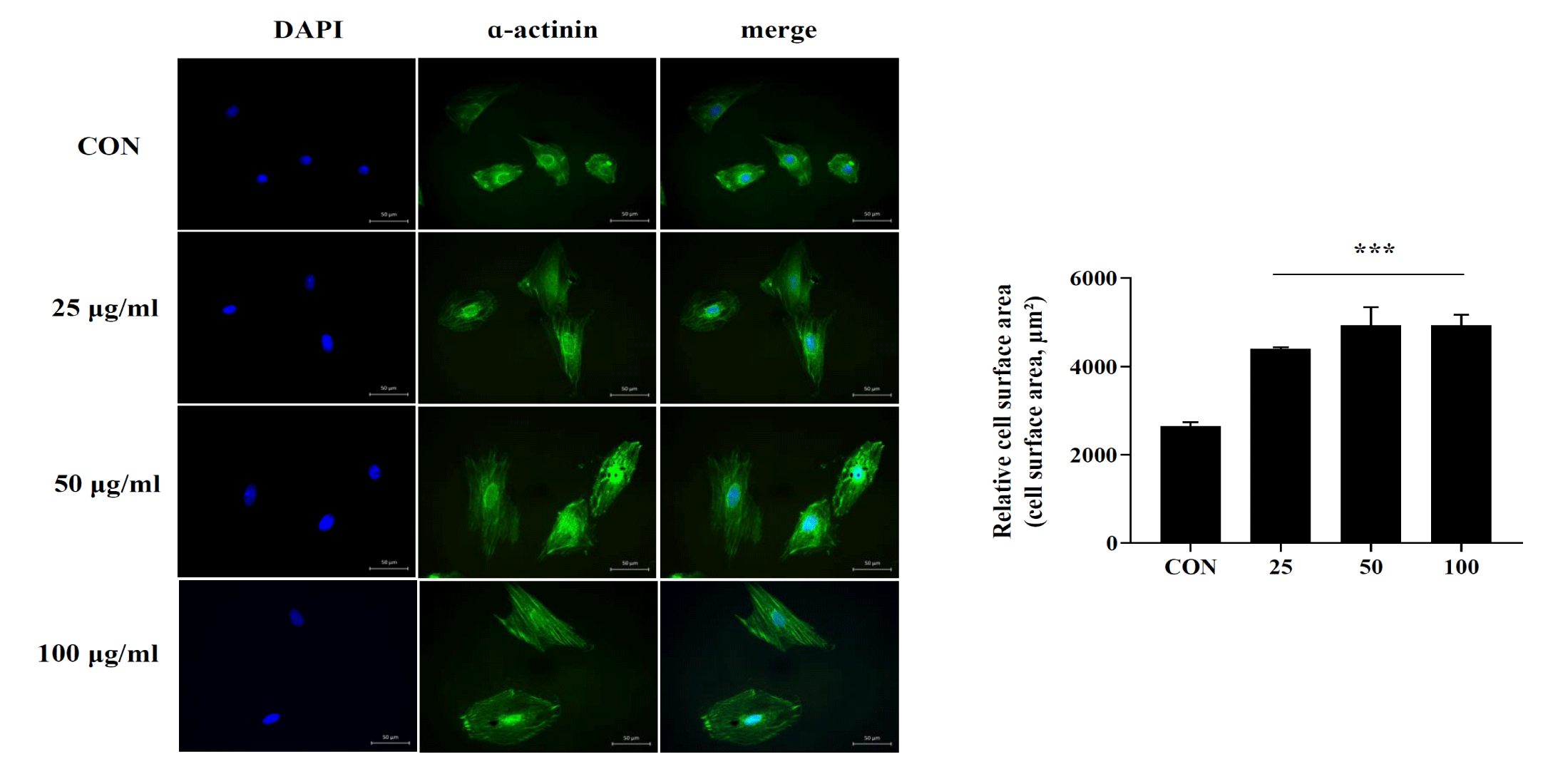
Cell surface area measurement
Myocardial disease is a physiological response of the heart to external and internal stimuli. Various pathophysiological stimuli promote a complex remodeling process in the heart, accompanied by cardiomyocyte loss, existing cardiomyocytes cell size enlargement, and cardiac hypertrophy, leading to ischemic and hypertrophic phenotypes [18]. Therefore, we investigated whether DPM could induce myocardial hypertrophy in vitro. H9C2 cells were treated with different concentrations of DPM for 3h, followed by immunostaining with α-actinin to measure cell size, as previously described. Our results showed that the DPM treatment group had markedly increased cell size compared to that in the control group (Figure 6).
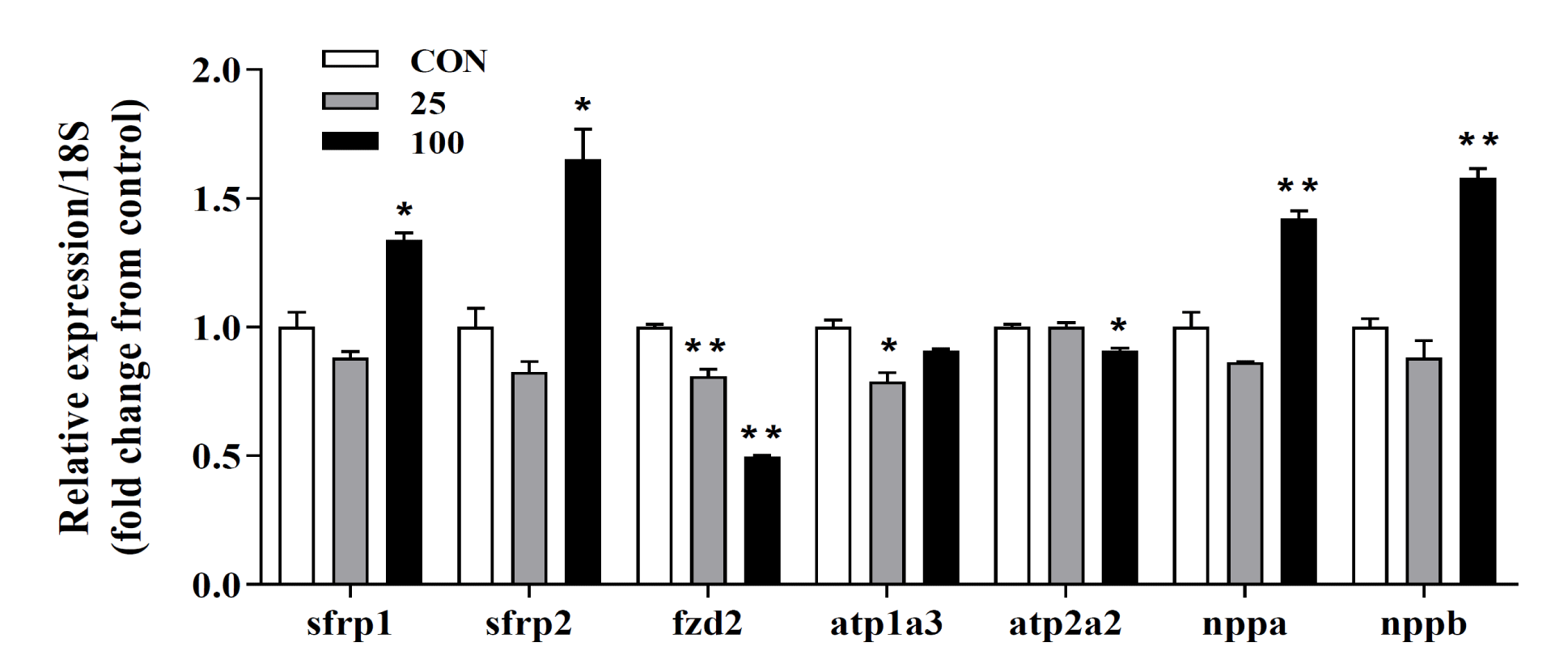
Validation of DEGs data using qRT-PCR
To verify the transcriptome data acquired after treating H9C2 cells with DPM for 24h, we used qRT-PCR using RNA extracted from stimulated cells. Among the analyzed transcripts, sfrp1, sfrp2, fzd2, and nppa showed similar expression profiles (Figure 7). DPM treatment appears to partially affect the Wnt-signaling pathway. The increased expression of fetal genes, such as natriuretic peptide precursor A (nppa) and natriuretic peptide precursor B (nppb), is associated with pathological hypertrophy [19,20]. We also investigated the expression levels of fetal gene mRNAs using qRT-PCR. There were significant increases in fetal gene expression at the transcriptional level, suggesting that there are mechanisms by which DPM facilitates myocardial hypertrophy (Figure 7).
Conclusion
We investigated the biological relationship between DPM and myocardial infarction based on the GEO database and bioinformatics. A total of 33 differently expressed genes were identified and their possible signaling networks were investigated. Furthermore, we revealed the activation of two cellular signaling pathways. In addition to increasing the prediction accuracy, the use of experimental validation and literature-based analysis in our study aids in understanding the mechanisms underlying DPM-related cardiac hypertrophy. The study results will be useful as a basis for future in-depth investigations.
Acknowledgment
Author contribution
This version of the article has been read and approved by all authors.
Funding
Not applicable.
Conflict of interest
The authors declare no conflict of interest.
References
- Giamouzis G, Dimos A, Xanthopoulos A, Skoularigis J, Triposkiadis F. Left ventricular hypertrophy and sudden cardiac death. Heart Fail Rev. 2022 Mar;27(2):711-724. doi: 10.1007/s10741-021-10134-5. Epub 2021 Jun 28. PMID: 34184173.
- Kang YJ. Cardiac hypertrophy: a risk factor for QT-prolongation and cardiac sudden death. Toxicol Pathol. 2006;34(1):58-66. doi: 10.1080/01926230500419421. PMID: 16507545.
- Shenasa M, Shenasa H. Hypertension, left ventricular hypertrophy, and sudden cardiac death. Int J Cardiol. 2017 Jun 15;237:60-63. doi: 10.1016/j.ijcard.2017.03.002. Epub 2017 Mar 3. PMID: 28285801.
- Liu R, Molkentin JD. Regulation of cardiac hypertrophy and remodeling through the dual-specificity MAPK phosphatases (DUSPs). J Mol Cell Cardiol. 2016 Dec;101:44-49. doi: 10.1016/j.yjmcc.2016.08.018. Epub 2016 Aug 27. PMID: 27575022; PMCID: PMC5154921.
- Zhang QJ, Tran TAT, Wang M, Ranek MJ, Kokkonen-Simon KM, Gao J, Luo X, Tan W, Kyrychenko V, Liao L, Xu J, Hill JA, Olson EN, Kass DA, Martinez ED, Liu ZP. Histone lysine dimethyl-demethylase KDM3A controls pathological cardiac hypertrophy and fibrosis. Nat Commun. 2018 Dec 7;9(1):5230. doi: 10.1038/s41467-018-07173-2. PMID: 30531796; PMCID: PMC6286331.
- MacDonald MR, Petrie MC, Hawkins NM, Petrie JR, Fisher M, McKelvie R, Aguilar D, Krum H, McMurray JJ. Diabetes, left ventricular systolic dysfunction, and chronic heart failure. Eur Heart J. 2008 May;29(10):1224-40. doi: 10.1093/eurheartj/ehn156. Epub 2008 Apr 18. PMID: 18424786.
- McEntee JC, Ogneva-Himmelberger Y. Diesel particulate matter, lung cancer, and asthma incidences along major traffic corridors in MA, USA: A GIS analysis. Health Place. 2008 Dec;14(4):817-28. doi: 10.1016/j.healthplace.2008.01.002. Epub 2008 Jan 11. PMID: 18280198..
- Gamble J. Lung cancer and diesel exhaust: a critical review of the occupational epidemiology literature. Crit Rev Toxicol. 2010 Mar;40(3):189-244. doi: 10.3109/10408440903352818. PMID: 20156057.
- Benbrahim-Tallaa L, Baan RA, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, Guha N, Loomis D, Straif K; International Agency for Research on Cancer Monograph Working Group. Carcinogenicity of diesel-engine and gasoline-engine exhausts and some nitroarenes. Lancet Oncol. 2012 Jul;13(7):663-4. doi: 10.1016/s1470-2045(12)70280-2. PMID: 22946126.
- Yan YH, Chen TB, Yang CP, Tsai IJ, Yu HL, Wu YS, Huang WJ, Tseng ST, Peng TY, Chou EP. Long-term exposure to particulate matter was associated with increased dementia risk using both traditional approaches and novel machine learning methods. Sci Rep. 2022 Oct 12;12(1):17130. doi: 10.1038/s41598-022-22100-8. PMID: 36224306; PMCID: PMC9556552.
- Hamanaka RB, Mutlu GM. Particulate Matter Air Pollution: Effects on the Cardiovascular System. Front Endocrinol (Lausanne). 2018 Nov 16;9:680. doi: 10.3389/fendo.2018.00680. PMID: 30505291; PMCID: PMC6250783.
- Du Y, Xu X, Chu M, Guo Y, Wang J. Air particulate matter and cardiovascular disease: the epidemiological, biomedical and clinical evidence. J Thorac Dis. 2016 Jan;8(1):E8-E19. doi: 10.3978/j.issn.2072-1439.2015.11.37. PMID: 26904258; PMCID: PMC4740122.
- Costello S, Attfield MD, Lubin JH, Neophytou AM, Blair A, Brown DM, Stewart PA, Vermeulen R, Eisen EA, Silverman DT. Ischemic Heart Disease Mortality and Diesel Exhaust and Respirable Dust Exposure in the Diesel Exhaust in Miners Study. Am J Epidemiol. 2018 Dec 1;187(12):2623-2632. doi: 10.1093/aje/kwy182. PMID: 30137203; PMCID: PMC6269247.
- Torén K, Bergdahl IA, Nilsson T, Järvholm B. Occupational exposure to particulate air pollution and mortality due to ischaemic heart disease and cerebrovascular disease. Occup Environ Med. 2007 Aug;64(8):515-9. doi: 10.1136/oem.2006.029488. Epub 2007 Feb 15. PMID: 17303673; PMCID: PMC2078490.
- Zuo L, Youtz DJ, Wold LE. Particulate matter exposure exacerbates high glucose-induced cardiomyocyte dysfunction through ROS generation. PLoS One. 2011;6(8):e23116. doi: 10.1371/journal.pone.0023116. Epub 2011 Aug 5. PMID: 21850256; PMCID: PMC3151271..
- Cao Y, Long J, Ji Y, Chen G, Shen Y, Gong Y, Li J. Foam cell formation by particulate matter (PM) exposure: a review. Inhal Toxicol. 2016 Nov;28(13):583-590. doi: 10.1080/08958378.2016.1236157. Epub 2016 Oct 6. PMID: 27706953.
- Park JH, Nho KJ, Lee JY, Yoo YJ, Park WJ, Cho C, Kim DH. Anti-Ischemic Effects of PIK3IP1 Are Mediated through Its Interactions with the ETA-PI3Kγ-AKT Axis. Cells. 2022 Jul 11;11(14):2162. doi: 10.3390/cells11142162. PMID: 35883611; PMCID: PMC9322903.
- Burchfield JS, Xie M, Hill JA. Pathological ventricular remodeling: mechanisms: part 1 of 2. Circulation. 2013 Jul 23;128(4):388-400. doi: 10.1161/CIRCULATIONAHA.113.001878. PMID: 23877061; PMCID: PMC3801217.
- Izumo S, Nadal-Ginard B, Mahdavi V. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc Natl Acad Sci U S A. 1988 Jan;85(2):339-43. doi: 10.1073/pnas.85.2.339. PMID: 2963328; PMCID: PMC279543.
- Song HK, Hong SE, Kim T, Kim DH. Deep RNA sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PLoS One. 2012;7(4):e35552. doi: 10.1371/journal.pone.0035552. Epub 2012 Apr 16. PMID: 22523601; PMCID: PMC3327670.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.