Biology Group . 2022 May 29;3(5):660-663. doi: 10.37871/jbres1492.
Four N-Genes in Using RT-PCR to Confirmed Coronavirus Disease-2019 (COVID-19) Pneumonia
Yao-Feng Hu1, Hao-Yu Liu2* and Te-Lung Tsai3
2Department of Medical Examiner, Hsinchu Mackay Memorial Hospital, 30071 Taiwan
3Department of Dean's Office Director, Hsinchu Mackay Memorial Hospital, 30071 Taiwan
- COVID-19
- SARS-CoV-2
- RT-PCR
- CPs
Abstract
The complete laboratory data of 24 samples with confirmed coronavirus Disease-2019 were retrieved by nucleic acid testing, and the quantification of SARS-CoV-2 with the World Health Organization (WHO) SARS-CoV-2 envelope (E-gene), RNA-Dependent RNA Polymerase (RdRP-gene), Nucleocapsid (N-gene) three primers by the Roche Cobas z 480. Throat or nasopharyngeal swabs reports demonstrated three primer Cross Points (CPs) of greater than 29, but less than 29 in sputum. COVID-19 confirmed patients demonstrated three genes E-, RdRp- and N-genes, actually four of 24 no showed N-gene, highly suspected to be weakly positive.
Introduction
The four common human coronaviruses; 229E, NL63, OC43, and HKU1, cause mild illnesses, such as the common cold. The 2003 Severe Acute Respiratory Syndrome (SARS) pandemic and the Middle East respiratory syndrome virus originating from Arabia brought this virus family into the public consciousness worldwide [1]. The 2019 Novel Coronavirus (2019-nCoV) was first reported at the end of December 2019 after dozens of visitors to a seafood market developed severe pneumonia (Coronavirus Disease 2019 (COVID-19)). The genome published on Jan. 11th (GenBank acc. MN908947) shows high similarity to the SARS virus, thus, the new name for the virus became SARS Coronavirus-2 (SARS-CoV-2). Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR) tests have been widely used to detect SARS-CoV-2 [2-5]. From March 13, 2020 to April 15, 2021, 9756 throat or nasopharyngeal swabs and sputum specimens were collected, 24 of which showed positive RT-PCR results. So far, the number of people diagnosed with COVID-19 in Taiwan is relatively stable and low compared to the rest of the world, but it still cannot be taken lightly. We followed the Taiwan CDC guidelines for using the real-time RT-PCR molecular diagnosis method, extracting RNA with a LabTurbo 24 Compact System (Taiwan) and undertaking quantitative PCR (qPCR) using a Cobas z 480 (Rotkreuz, Switzerland). For rapid point-of-care tests, few have been accredited by the CDC and checked and registered by the Taiwan Food and Drug Administration (TFDA), and these only have TFDA Emergency Use Authorization (EUA). This study involving positive RT-PCR reports to the Taiwan CDC confirmed their use as a successful diagnosis platform, indicating that patients who test positive for COVID-19 should be placed in negative pressure wards as quickly as possible.
Materials and Methods
Subjects
Twenty patients with COVID-19 infection inspected by 24 throat or nasopharyngeal swabs and sputum COVID-19 with Roche Cobas z 480 analyzer nucleic acid tests from 13 March, 2020 to 15 April, 2021 at Hsinchu Mackay Memorial Hospital were included in this study. All the patients were treated according to the “New Coronavirus (SARS-CoV-2) Interim Guidelines for Clinical Treatment of Infections” announced by the Taiwan CDC [6].
Analysers and reagents
The swabs and sputum were mixed with 1 ml of Dulbecco’s modified Eagle’s medium (Gibco BRL, Grand Island, NY, USA) and the sputum samples were vortexed and mixed well with phosphate-buffered saline. Total viral RNA was extracted using a LabPrep Viral DNA/RNA Mini Kit by LabTurbo 24 Compact System (LabTurbo 24C). Overall, 5 μl of total RNA was subjected to analysis of the COVID-19 gene by Tib-Molbiol's 2019-nCoV Real-Time RT-PCR kit. For the quantification of SARS-CoV-2 with the World Health Organization (WHO) SARS-CoV-2 envelope (E-gene), RNA-Dependent RNA Polymerase (RdRP-gene), nucleocapsid (N-gene), and Equine Arteritis Virus (EAV) extraction control targets, such as real-time PCR with LightCycler® Multiplex RNA Virus Master and Wuhan COVID-19-specific primer with probe (all from TIB MolBiol, Cat.-No. 53-0776-96), were subjected to RNA extraction control with EAV (TIB MolBiol, Cat.-No. 66-0909-96). All real-time PCR assays were performed on a Roche Cobas z 480 Analyzer, and the values were analyzed according to Lightcycler® 480 manufacturer specifications.
Principles of analysers
Nucleic acid was obtained from patient samples by the addition of proteinase K and sample lysis thermology reagent in a class Ⅱ biological safety cabinet with a negative pressure room containing mycobacterial culture, with some laboratories at biosafety level three, using a fully automatic column extraction machine for approximately 1 h 20 min. Selective amplification of target nucleic acids from a sample was achieved with target-specific forward and reverse primers for open reading frame 1, a nonstructural region that is unique to SARS-CoV-2 [7]. Additionally, a conserved region in the structural protein envelope E-gene was selected for pan-Sarbecovirus detection, which can also be used to detect the SARS-CoV-2 virus. Selective amplification of RNA internal control was achieved by using noncompetitive sequence-specific forward and reverse primers that had no homology with the CoV genome.
A parameter-specific reagent (Tib-Molbiol) contained the specific primers and probes for each E-gene, RdRp gene, and N-gene. A 76 bps fragment from a conserved region in the E-gene was detected with 5-Carboxyfluorescein (FAM)-labeled hydrolysis probes. A 100 bps fragment from a conserved region of the RdRP gene was detected with a SARS-CoV-2-specific FAM-labeled hydrolysis probe. A 128 bps fragment from the N-gene was detected with FAM-labeled hydrolysis probes. This assay detects SARS and the 2019-nCoV pneumonia virus as well as other bat-associated SARS-related viruses (Sarbecovirus). The experiments were performed with a Roche Cobas z480. This assay detects SARS and the 2019-nCoV pneumonia virus as well as other bat-associated SARS-related viruses (Sarbecovirus) [8].
Results
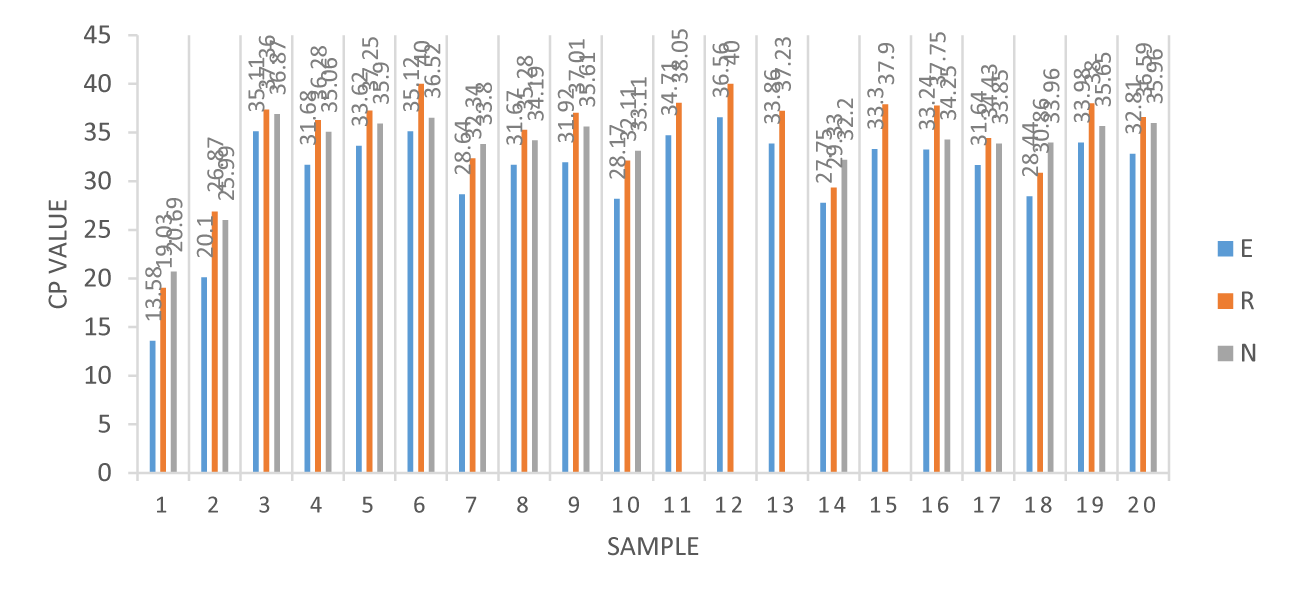
The samples were obtained from 24 throat or nasopharyngeal swabs and sputum; swabs reports demonstrated three primer Cross Points (CPs) of greater than 29, but less than 29 in sputum, as shown in table 1 [9]. Twenty throat or nasopharyngeal swabs were analyzed for E-, RdRp-, and N-genes, which were found to have average means of 29.07, 33.22 and 33.35 Cross Points (CPs), respectively. However, four in sputum, the E-, RdRp-, and N-gene average means were 20.44, 25.92 and 25.26 Cross Points (CPs), respectively. The CPs values of 20 throat or nasopharyngeal swabs of COVID-19 were confirmed by real-time RT-PCR using blue, orange, and gray bar charts representing the E-, RdRp- and N-genes, respectively, as illustrated in figure 1. In theory, COVID-19 confirmed patients demonstrated three genes E-, RdRp- and N-genes, actually four of 20 showed no N-gene, highly suspected to be weakly positive.
| Table 1: Three primer Cross Points (CPs) of 22 diagnosed samples in the throat or nasopharyngeal swabs that indicated a value greater than 29 CPs and in sputum with a value of less than 29 CPs. | |||
| Throat or nasopharyngeal swabs | E-gene (CPs) | RdRp-gene(CPs) | N-gene (CPs) |
| 1 | 13.58 | 19.03 | 20.69 |
| 2 | 20.1 | 26.87 | 25.99 |
| 3 | 35.11 | 37.36 | 36.87 |
| 4 | 31.68 | 36.28 | 35.06 |
| 5 | 33.62 | 37.25 | 35.9 |
| 6 | 35.12 | 36.52 | 40 |
| 7 | 28.64 | 32.34 | 33.8 |
| 8 | 31.67 | 35.28 | 34.19 |
| 9 | 31.92 | 37.01 | 35.61 |
| 10 | 28.17 | 32.11 | 33.11 |
| 11 | 34.71 | 38.05 | X |
| 12 | 36.56 | 40 | X |
| 13 | 33.86 | 37.23 | X |
| 14 | 27.75 | 29.33 | 32.2 |
| 15 | 33.3 | 37.9 | X |
| 16 | 33.24 | 37.75 | 34.25 |
| 17 | 31.64 | 34.43 | 33.85 |
| 18 | 28.44 | 30.86 | 33.96 |
| 19 | 33.98 | 38 | 35.65 |
| 20 | 32.81 | 36.59 | 35.96 |
| Mean | 29.07 | 33.22 | 33.35 |
| Sputum | E-gene (CP) | RdRp-gene (CPs) | N-gene (CPs) |
| 1 | 12.63 | 18.82 | 19.02 |
| 2 | 22.51 | 27.63 | 29.14 |
| 3 | 24.78 | 31.8 | 27.86 |
| 4 | 21.82 | 25.43 | 29.03 |
| Mean | 20.44 | 25.92 | 25.26 |
Discussion
The global epidemic of COVID-19 has shown an expanding trend for example, number of cases or range of transmission again as of late June. From March 13, 2020 to April 15, 2021, twenty-four of 9,756 throat or nasopharyngeal swabs and sputum were eventually confirmed to have COVID-19 by RT-PCR of including one retrospective inspection, positive ratio only up to 0.24 % in our clinical samples, might exist a potentially false negative rate of RT-PCR testing for SARS-CoV-2 [10]. This might be caused by insufficient viral material or proper respiratory tract specimens at the right time, laboratory mistakes during sampling, or restrictions on sample transportation. Some recent reviews have demonstrated high viral loads in the respiratory tract within five to six days of symptoms, nasopharyngeal swab positive ratios more than throat swabs, so early screening may be necessary [7]. Furthermore, the data indicated that sputum CPs were fewer than those of nasopharyngeal or throat swabs, indicating high viral loads in the lower respiratory tract compared to in the upper tract [9]. Using real-time PCR, amplification and analysis were performed in a closed system to minimize false-positive reports due to contamination. Limited real-time RT-PCR assays should not be used to quantify COVID-19 viral loads. The 20 SARS-CoV-2 positive patients were confirmed by RT-PCR to demonstrate three primer CPs, including E-, RdRp-, and N-genes. According to real-time RT-PCR CPs values of COVID-19 pneumonia, the sample 11 to 13 and 15 that indicate no N-gene, which could be the CPs of the E-gene, the RdRp-gene was closer to 40, and reasonable suspicion was weak positive SARS-CoV-2. Through the Taiwan CDC confirmed an infection of COVID-19, previous infection titers may fall, and this result appeared in the family group contact infection involved in this study. After a year of Wuhan pneumonia in Taiwan, the prevention of overseas travel and implementation of social isolation and mask wearing occurred, as was the case for many countries globally. Fortunately, the number of confirmed cases in Taiwan has been isolated to small areas, but very few patients have died of nCoV pneumonia.
Ethical approval
This study was approved by Mackay Memorial Hospital Institutonal Review Broad Approval of Clinical Trial. (Approval number 20MMHIS184e).
Conflicts of interest
The authors have no conflicts of interest.
References
- Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020 Mar 15;16(10):1686-1697. doi: 10.7150/ijbs.45472. PMID: 32226286; PMCID: PMC7098031.
- Carter JG, Orueta Iturbe L, Duprey JHA, Carter IR, Southern CD, Rana M, Whalley CM, Bosworth A, Beggs AD, Hicks MR, Tucker JHR, Dafforn TR. Ultrarapid detection of SARS-CoV-2 RNA using a reverse transcription-free exponential amplification reaction, RTF-EXPAR. Proc Natl Acad Sci U S A. 2021 Aug 31;118(35):e2100347118. doi: 10.1073/pnas.2100347118. PMID: 34400545; PMCID: PMC8536344.
- Visseaux B, Le Hingrat Q, Collin G, Bouzid D, Lebourgeois S, Le Pluart D, Deconinck L, Lescure FX, Lucet JC, Bouadma L, Timsit JF, Descamps D, Yazdanpanah Y, Casalino E, Houhou-Fidouh N; Emergency Department Influenza Study Group. Evaluation of the QIAstat-Dx Respiratory SARS-CoV-2 Panel, the First Rapid Multiplex PCR Commercial Assay for SARS-CoV-2 Detection. J Clin Microbiol. 2020 Jul 23;58(8):e00630-20. doi: 10.1128/JCM.00630-20. PMID: 32341142; PMCID: PMC7383528.
- Mardian Y, Kosasih H, Karyana M, Neal A, Lau CY. Review of Current COVID-19 Diagnostics and Opportunities for Further Development. Front Med (Lausanne). 2021 May 7;8:615099. doi: 10.3389/fmed.2021.615099. PMID: 34026773; PMCID: PMC8138031.
- Poljak M, Korva M, Knap Gašper N, Fujs Komloš K, Sagadin M, Uršič T, Avšič Županc T, Petrovec M. Clinical Evaluation of the cobas SARS-CoV-2 Test and a Diagnostic Platform Switch during 48 Hours in the Midst of the COVID-19 Pandemic. J Clin Microbiol. 2020 May 26;58(6):e00599-20. doi: 10.1128/JCM.00599-20. PMID: 32277022; PMCID: PMC7269406.
- Recommendations for COVID-19: Case definition, specimen collection, and Diagnostic Tests. April, 2020.
- Tang YW, Schmitz JE, Persing DH, Stratton CW. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J Clin Microbiol. 2020 May 26;58(6):e00512-20. doi: 10.1128/JCM.00512-20. PMID: 32245835; PMCID: PMC7269383.
- Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin Chem. 2020 Apr 1;66(4):549-555. doi: 10.1093/clinchem/hvaa029. PMID: 32031583; PMCID: PMC7108203.
- Wang X, Tan L, Wang X, Liu W, Lu Y, Cheng L, Sun Z. Comparison of nasopharyngeal and oropharyngeal swabs for SARS-CoV-2 detection in 353 patients received tests with both specimens simultaneously. Int J Infect Dis. 2020 May;94:107-109. doi: 10.1016/j.ijid.2020.04.023. Epub 2020 Apr 18. PMID: 32315809; PMCID: PMC7166099.
- Li Y, Yao L, Li J, Chen L, Song Y, Cai Z, Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020 Jul;92(7):903-908. doi: 10.1002/jmv.25786. Epub 2020 Apr 5. PMID: 32219885; PMCID: PMC7228231.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.