Biology Group 2025 January 31;6(1):082-085. doi: 10.37871/jbres2058.
Detection of an Imported Case of Severe Crimean-Congo Hemorrhagic Fever in a Patient with Comorbidities, Dakar, Senegal 2023
Samba Niang Sagne1*, Ousseynou Sene2, Idrissa Dieng2, Mamadou Korka Diallo1, Amadou Moustapha Ndoye1, Moufid Mhamadi2, Safietou Sankhe2, Yoro Sall3, Boly Diop3, Oumar Faye2, Abdourahmane Sow4, Boubacar Diallo4, Moussa Moise Diagne2, Cheikh Loucoubar1, Gamou Fall2 and Mamadou Aliou Barry1
2Pole of Virology, Institut Pasteur de Dakar, Dakar, Senegal
3Prevention Department Ministry of Health, Dakar, Senegal
4Public Health Department, Institut Pasteur de Dakar, Dakar, Senegal
- Imported
- Severe
- Crimean-Congo hemorrhagic fever
- Comorbidities
- Senegal 2023
Abstract
In July 2023, a diabetic from Mauritania was diagnosed as a severe case of Crimean-Congo hemorrhagic fever (CCHF) at Dakar hospital, Senegal. The phylogenetic analysis revealed the new strain as a CCHF virus reassortant between Genotype Ⅰ and Ⅲ, closely linked to strains from Spain, Mauritania, Senegal and South Africa. Genetic variability of the virus in West Africa underscores the urgent need for enhanced surveillance in West Africa.
Introduction
Crimean-Congo Hemorrhagic Fever (CCHF) have been reported across Africa, Asia and Europe regions where the primary vectors Hyalomma ticks are prevalent. CCHF infection is correlated with several risk factors, including the bites of ticks, the contact with a patient with CCHF during the acute phase of the infection as well as with blood or tissues from viremic livestock [1]. The initial symptoms of CCHF are nonspecific and can advance to a hemorrhagic phase, with fatality rate ranging from 20 to 30% [2].
The virus CCHFV belongs to the Orthonairovirus genus of the Nairoviridae family characterized by a negative tripartite RNA genome with three segments namely S (Small), M (Medium), and L (Large) [3]. The current CCHFV classification includes five genotypes: Ⅰ-Ⅲ in Africa, Ⅳ in Asia and Ⅴ in Europe [4]. In West Africa, the first human CCHF cases were reported in Mauritania in 1983 [5]. Since them numerous outbreaks were described in Mauritania while sporadic cases were identified in Senegal [5-7]. Here, we report the longitudinal clinical and biological observations of a CCHF case imported from Mauritania to Senegal in 2023 as well as the genomic characterization of the detected virus.
The Study
In July 2023, a 59-year-old man with type 2 diabetes was admitted to emergency in Dakar hospital for febrile syndrome with negative rapid malaria test, an algic syndrome, an anemic syndrome, diffuse petechiae and purpura, vomiting and asthenia. The illness had begun 02 weeks prior to admission. Biological results revealed severe hyperglycemia at 4.86 g/dl with glycosuria and absence of ketonuria, negative thickened gout, C-reactive protein at 28 mg/l, hemoglobin at 9.7 g/dl, thrombocytopenia at 2000/mm3, impaired renal function creatinemia at 46 mg/l, hyponatremia at 131 mEq/l, hypochloremia at 91 mEq/l, hepatic cytolysis with SGOT transaminase at 2020 IU/l, SGPT transaminase at 762 IU/l, biological cholestasis, direct bilirubin increased to 9.7 mg/l. He was treated with antipyretics, insulinotherapy and platelet concentrate transfusions. The evolution marked by the occurrence of gingivorrhagia and epitaxis. A Viral Hemorrhagic Fever (VHF) suspected, blood sample taken and sent to Institut Pasteur Dakar (IPD) laboratory. He succumbed two days after his admission from hemorrhagic shock. At IPD, viral RNA was extracted from the clinical sample using the QIAamp viral RNA mini kit according to the manufacturer's instructions (Qiagen, Hilden, Germany) and screened for several arboviruses and VHFs including CCHFV by real time RT-PCR [8]. The results confirmed the presence of CCHFV in the sample. Whole genome sequencing was performed following an hybrid capture approach with the Twist Biosciences Comprehensive Viral Research Panel (CVRP) as previously described on an Illumina iSeq100 instrument [9]. The sequencing reads were analyzed using the open-source metagenomics CZ-ID platform (http://czid.org), employing default threshold filters for quality checking reads, base-calling and generating consensus sequences. Nearly complete genomes obtained during this work were submitted to a public nucleotide BLAST database to identify their homologous sequences. Sequences were aligned using the MAFFT program and Maximum Likelihood phylogenetic tree build with IQ-TREE with 1000 bootstrap replicates for robustness [10,11]. Our study reports a CCHF case imported from Mauritania in a patient with comorbidities (diabetic). Regrettably, the patient succumbed to the complication. This case demonstrates clinical and biological similarities with the initial documented CCHF cases in West Africa including the first instance in Mauritania and the 2004 case involving two tourist in Dakar in 2004 [6,7].
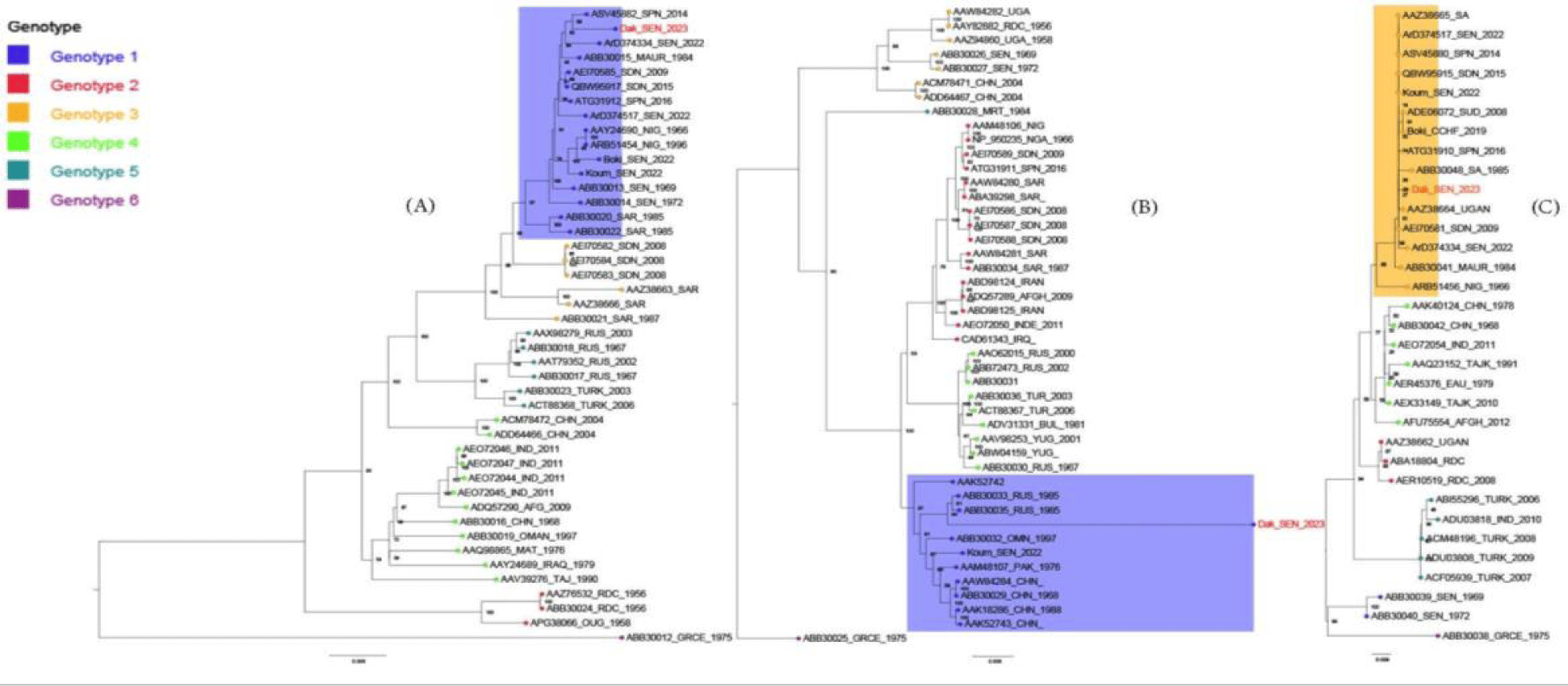
BLASTn analyses revealed that the newly identified strain shares over 98% nucleotide sequence similarity with strains isolated from Spain in 2014 and 2016 (ASV45882, ATG31912), Senegal in 1975 (WAD86875), Mauritania in 1984 (ABB30041) and Sudan in 2009 (AEI70581). Phylogenetic analysis revealed that the new CCHF strain is a probable reassortant between Genotype I (L and M segment) and Ⅲ (S segment) and clustered with the recent known reassortant strain from Koumpentoum, Eastern Senegal in 2022 (Koum_SEN_2022) along the three segment [12]. Furthermore, these strains appear to have multiples origins as they form clusters with strain from Spain in 2014 and 2016, Mauritania in 1984, Senegal in 2019 and 2022 (ArD374334, Boki_CCHF_2019) and South-Africa in 1985 (AAZ38665, ABB30048) across the L, M and S segments (Figure 1). Reassortment in bunyaviruses can alter their pathogenicity, exemplified by Ngari virus, a reassortant orthobunyavirus with L and S segment from Bunyamwera virus and the M segment from Batai virus. Although Bunyamwera and Batai viruses typically cause mild disease, Ngari has been associated with hemorrhagic fever cases in East Africa [13,14] . Therefore, it is crucial to investigate the potential impact of this reassortment on the biological properties of the new strains and ascertain its potential involvement in the observed severe clinical manifestations. The patient travelled from Mauritania, an epidemic country for CCHFV, supporting the idea of transboundary transmission and strain exchange between Senegal and Mauritania.
Our study highlights Mauritania’s high-risk status for CCHF and confirm the hypothesis of strains exchange between different countries including Senegal and Mauritania. Additionally, it sheds light on the genetic diversity among West African CCHF strains and raises question about the potential introduction of CCHF strains from other countries including Spain and South Africa and underscoring the need for enhanced surveillance to prevent future outbreaks. It also emphasizes the importance of considering comorbidity factors, such as diabetes, in the management of CCHF cases. Lack of early detection of CCHF in Mauritania might be due to the unspecific symptoms during the acute stage of the disease or to inefficient surveillance system.
Acknowledgement
We thank Cherif Sylla, Diogop Camara, Maïmouna Mbanne and other staff at the WHO CC for arbovirus and VHF for their technical assistance.
Funding
This work was supported by the Institut Pasteur de Dakar proper funds, Senegal.
Disclosure Statement
The authors declare no conflicts of interest.
References
- Ergonul O. Crimean-Congo hemorrhagic fever virus: new outbreaks, new discoveries. Curr Opin Virol. 2012 Apr;2(2):215-20. doi: 10.1016/j.coviro.2012.03.001. PMID: 22482717.
- Whitehouse CA. Crimean-Congo hemorrhagic fever. Antiviral Res. 2004 Dec;64(3):145-60. doi: 10.1016/j.antiviral.2004.08.001. PMID: 15550268.
- Mazzola LT, Kelly-Cirino C. Diagnostic tests for Crimean-Congo haemorrhagic fever: a widespread tickborne disease. BMJ Glob Health. 2019 Feb 20;4(Suppl 2):e001114. doi: 10.1136/bmjgh-2018-001114. PMID: 30899574; PMCID: PMC6407549.
- Shayan S, Bokaean M, Shahrivar MR, Chinikar S. Crimean-Congo Hemorrhagic Fever. Lab Med. 2015 Summer;46(3):180-9. doi: 10.1309/LMN1P2FRZ7BKZSCO. PMID: 26199256.
- Camicas JL, Gonzalez JP, Leguenno B, Paul CJ, Digoutte JP, Calvo MA, François A, Wilson ML. Recherches menées conjointement par l’USAMRIID, L’IPD et l’ORSTOM sur l’épidémiologie de la fièvre hémorragique de Crimée Congo au Sénégal et en Mauritanie. 1989.
- Tall A, Sall AA, Faye O, Diatta B, Sylla R, Faye J, Faye P, Ly AB, Sarr FD, Diab H, Diallo M. Deux cas de fièvre hémorragique de Crimée-Congo (FHCC) contractée au Sénégal, en 2004, par des résidentes temporaires. Bull Soc Pathol Exot. 2009.
- Saluzzo JF, Aubry P, Aubert H, Digoutte JP. La maladie à virus CHF-Congo en Afrique. A propos d'un cas à manifestations hémorragiques en Mauritanie [Crimean-Congo hemorrhagic fever in Africa. Apropos of a case with hemorrhagic manifestations in Mauritania]. Bull Soc Pathol Exot Filiales. 1985;78(2):164-9. French. PMID: 4028308.
- Weidmann M, Faye O, Faye O, Abd El Wahed A, Patel P, Batejat C, Manugerra JC, Adjami A, Niedrig M, Hufert FT, Sall AA. Development of Mobile Laboratory for Viral Hemorrhagic Fever Detection in Africa. J Infect Dis. 2018 Oct 5;218(10):1622-1630. doi: 10.1093/infdis/jiy362. PMID: 29917112; PMCID: PMC6173574.
- Crispell G, Williams K, Zielinski E, Iwami A, Homas Z, Thomas K. Method comparison for Japanese encephalitis virus detection in samples collected from the Indo-Pacific region. Front Public Health. 2022 Nov 24;10:1051754. doi: 10.3389/fpubh.2022.1051754. PMID: 36504937; PMCID: PMC9730272.
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002 Jul 15;30(14):3059-66. doi: 10.1093/nar/gkf436. PMID: 12136088; PMCID: PMC135756.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015 Jan;32(1):268-74. doi: 10.1093/molbev/msu300. Epub 2014 Nov 3. PMID: 25371430; PMCID: PMC4271533.
- Sene O, Sagne SN, Ngom D, Diagne MM, Badji A, Khoulé A, Ndiaye El H, Sankhe S, Loucoubar C, Diallo M, Weidmann M, Dia N, Simon-Lorière E, Sall Y, Diop B, Ndiaye M, Sakuntabhai A, Sall AA, Faye O, Faye O, Diallo D, Barry MA, Fall G. Emergence of crimean–congo hemorrhagic fever virus in eastern senegal in 2022. Viruses. 2024;16(2):315. doi: 10.3390/v16020315.
- Sall AA, Zanotto PM, Sene OK, Zeller HG, Digoutte JP, Thiongane Y, Bouloy M. Genetic reassortment of Rift Valley fever virus in nature. J Virol. 1999 Oct;73(10):8196-200. doi: 10.1128/JVI.73.10.8196-8200.1999. PMID: 10482570; PMCID: PMC112837.
- Bowen MD, Trappier SG, Sanchez AJ, Meyer RF, Goldsmith CS, Zaki SR, Dunster LM, Peters CJ, Ksiazek TG, Nichol ST; RVF Task Force. A reassortant bunyavirus isolated from acute hemorrhagic fever cases in Kenya and Somalia. Virology. 2001 Dec 20;291(2):185-90. doi: 10.1006/viro.2001.1201. PMID: 11878887.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.