Biology Group . 2022 December 31;3(12):1565-1566. doi: 10.37871/jbres1636.
Introduction to One Drop of Body Fluid - PCR (1DF-PCR)
Kuo-Ping Chiu*
It is an attractive idea to make a single drop of blood sufficient for the diagnosis of many diseases. This was exactly what the biotech company Theranos Inc., founded in 2003 by a 19-year-old girl, Elizabeth Holmes, claimed. Unfortunately, their health technology turned out to be a fraud, and Elizabeth was charged by the U.S. Securities and Exchange Commission and federal government for business crime and wire fraud. However, people should not be discouraged by the negative story. As science inches forward, biotechnologies will become more sophisticated, making materials to be used more efficiently and effectively.
Here, I introduce the 1DF-PCR (one drop of body fluid - Polymerase Chain Reaction) technology which allows us to produce sufficient molecules from a minute DNA/cDNA sample, such as cell-free DNA (cfDNA) in body fluids or forensic DNA sample, to be used in a number of purposes including sequencing, noninvasive prenatal testing (NIPT), forensic STR (short tandem repeat) investigation, etc.
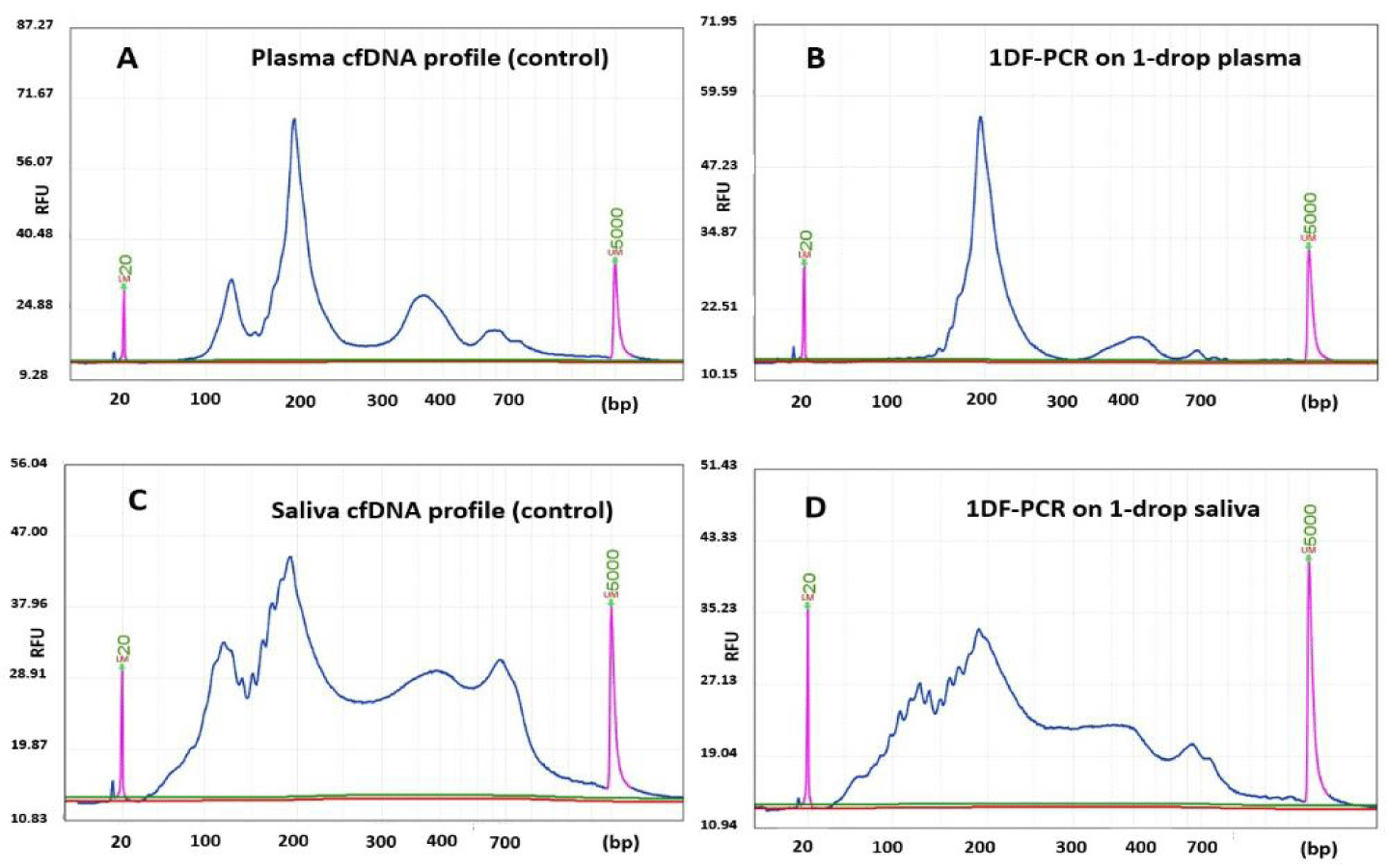
Cell-free nucleic acids (cfNAs) in liquid biopsies are important biomarkers and genetic materials for biomedical studies and diagnostics. However, normally a few milliliters of blood, or other body fluids, are required to obtain a sufficient quantity of cfNAs for multiple analyses. Moreover, most methods require cfNA to be isolated before experimentation. These prerequisites seriously hinder the progress of minute DNA analyses. 1DF-PCR is able to directly amplify the cfDNA fragments from 10 µL, or even less, of body fluid without purification (Figure 1). 1DF-PCR maintains a high degree of amplification fidelity and efficiency (e value >= 0.6). Unlike the method currently used in the sequencing industry, 1DF-PCR focuses solely on making the target cfDNA fragments without producing artificial, nonspecific products. As such, the original profile is well maintained. To the best of our knowledge, it is the most reliable adapter-dependent PCR method being able to directly and faithfully amplify cfDNA from a crude body fluid.
Potential interference by nonspecific substrates is empirically eliminated or at least minimized. Firstly, cells in the body fluid are eliminated by cold centrifugation (18,000 g for 10 min). Proteins and other macromolecules are subsequently heat denatured to alter their physical structures and biochemical functions. PH is largely maintained by PCR buffer, even after dilution. At same time, ions in body fluids do not seem to have detectable effect on amplification efficiency.
1DF-PCR has also been successfully tested for forensic samples. Under such circumstances, digestion of the sample with restriction enzymes is required because forensic samples mostly contain large DNA fragments such as chromosomes or large chromosomal fragments, which need to be made smaller for efficient amplification. Furthermore, when applied to quantitative analysis of a population of molecules such as transcriptome sequencing, gene copy number variations and microRNA expression (reverse transcription to cDNA required), although the method is able to amplify a single molecule, the quantity of liquid biopsy has to contain sufficient DNA molecules so to make the sample representative. We believe that the 1DF-PCR method should be feasible for recovering degraded DNA samples or DNA in formalin-fixed paraffin-embedded (FFPE) samples, although not yet empirically tested.
In summary, unlike that claimed by Theranos, 1DF-PCR is an evidence-based method. Here, we use it as an example to convince the biotech industry and the world that the idea is still worth pursuing and it will become even more prevalent in the future.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.