Medicine Group . 2022 September 07;3(9):1012-1015. doi: 10.37871/jbres1546.
A Neurodevelopmental Disorder with Intellectual Disability and Autism due to MAP7D3 Haploinsufficiency
Enrique Nogueira1-3*, Beatriz del Olmo1, Concepción Lobo1, Génesis Vizuete1 and Carmen Garma4
2Clinical Genetics Service, Hospital La Zarzuela, Madrid, Spain
3Clinical Genetics Service, Hospital San Rafael, Madrid, Spain
4Health Diagnostics, Quirónsalud, Madrid, Spain
- MAP7D3
- Haploinsufficiency
- Intellectual disability
- Autism
Summary
We report on a young boy with a neurodevelopmental disorder who is a carrier of a novel frameshift mutation in gene MAP7D3 (MAP7 domain containing 3) of Xq26.3. The protein encoded by this gene belongs to the MAP7 (microtubule associated protein 7) family that is proposed to regulate kinesin-1 (KIF5B)-dependent intracellular transport, by acting as Microtubule (MT)-tethered recruitment factors and activators of this kinesin [1-3]. And as other MT-associated proteins is conceivable the involvement of MAP7D3 in neuronal morphogenesis and therefore in neurodevelopmental disorders.
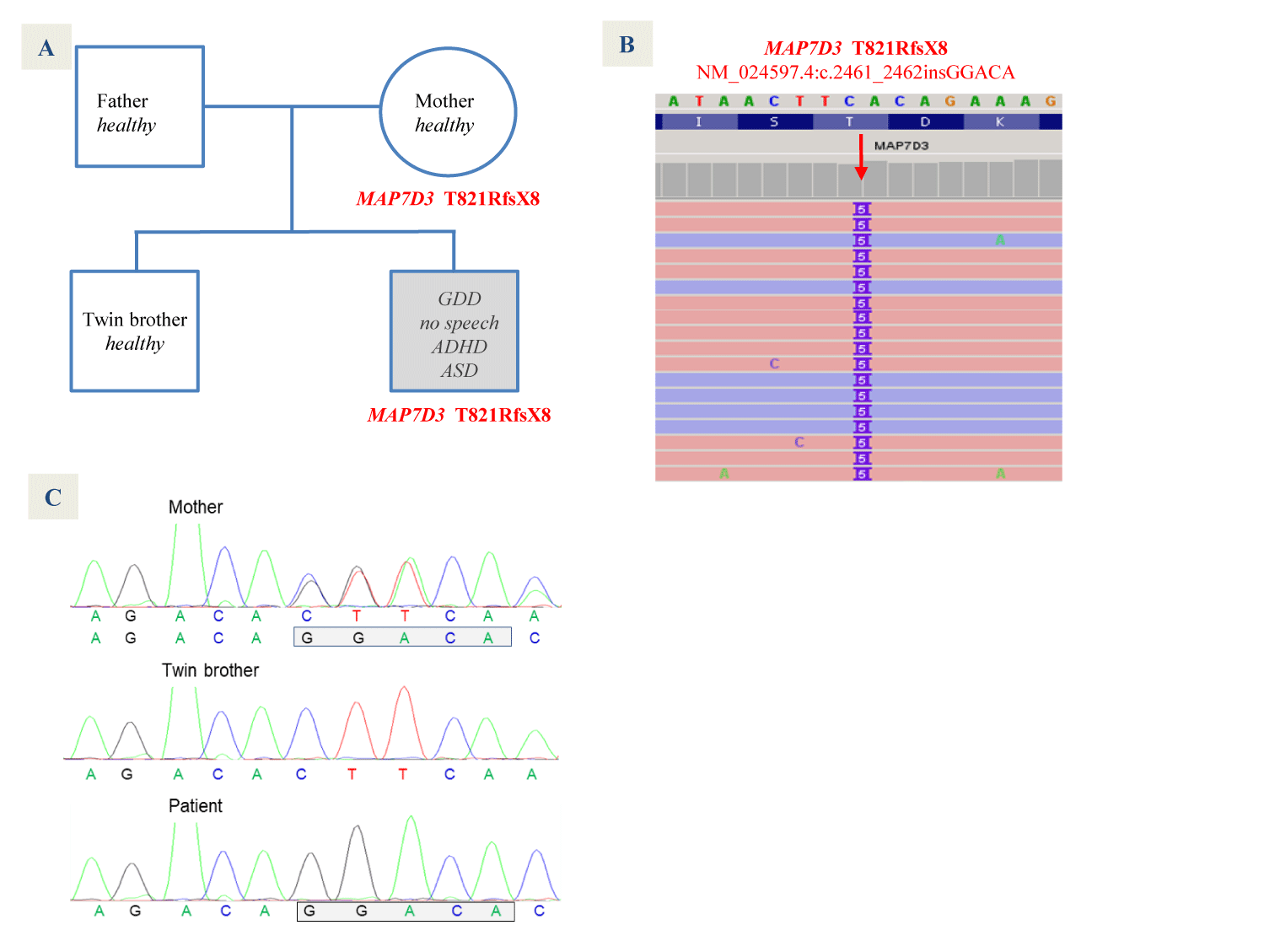
The findings described here arise from the genomic analysis of an 11-year-old boy under investigation for the last five years for a disorder with Global Developmental Delay (GDD), marked difficulty in language acquisition and learning, as well as Attention-Deficit/Hyperactivity and Autism Spectrum Disorders (ADHD, ASD). He lacks dysmorphic features and additional relevant findings, including those of chromosome studies (normal 46,XY karyotype), a genomic CGH-array (750 K), brain MRI, sleep EEG, and diverse clinical analyses, including that of 5'UTR CGGn sequence of gene FMR1 involved in most cases of fragile X syndrome, with normal results (a sequence of normal size, with 30 CGG triplets). He does not have a family history of interest, being a descendant of healthy fathers, like his twin brother.
Clinical exome analysis
It was undertaken with Illumina NGS (Next Generation Sequencing) procedures, initially (in the year 2018) with the TruSight One Sequencing Panel (for 4.813 genes) and later on (in year 2020) based on WES (Whole Exome Sequencing, with the use of IDT probes) as described in detail at Illumina, https://www.illumina.com. It resulted in the discovery in 2018 of frameshift novel mutation T821RfsX8 (p.Thr821ArgfsTer8) in gene MAP7D3, which is considered the only relevant genomic finding for the evaluation of patient disorder. No other mutations of interest, including exon deletions and duplications, were discovered with the evaluation of numerous genes with proposed association to cardinal features of the patient disorder according to HPO database (> 1700, > 900, > 340, and > 600 genes of GDD, delayed speech and language development, ADHD and ASD, respectively) submitted to WES variant analysis with use of two different pieces of software, of Variant Interpreter (Illumina) and Franklin (Genoox; https://franklin.genoox.com).
As it is depicted by figures 1A-C frameshift mutation T821RfsX8 is due to variant c.2461_2462insGGACA of exon 16 (in reference sequence NM_024597.4), present in a hemizygous pattern in all single sequences in the NGS BAM file of the patient (Figure 1B). The mutation is derived from a maternal allele not inherited by the twin brother (Figure 1C), who, like the mother and the father, is healthy.
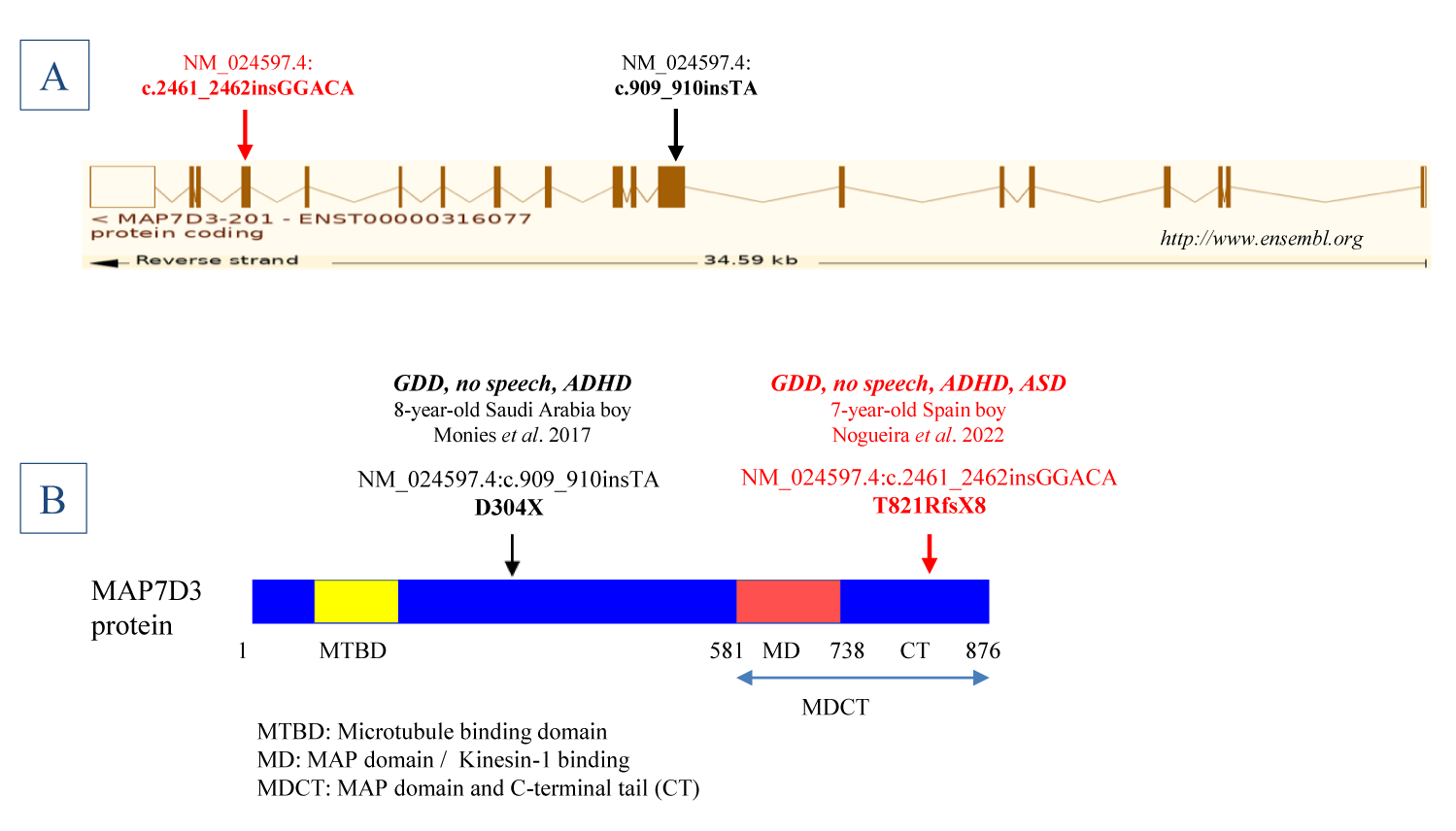
Further, and as it is shown by figures 2A & B T821RfsX8 mutation is located at the C-terminal end of MAP7D3 protein, far from exon 8 nonsense mutation D304X (p.Asp304Ter) due to variant c.909_910insTA (referred to sequence NM_024597.4) reported by Monies, et al. [4] as a finding of genetic diseases in Saudi Arabia based on the first 1000 diagnostic panels and exomes. It was identified in an 8-year-old boy, a descendant of non-consanguineous healthy parents, affected by GDD, ADHD, no speech, and mild hepatosplenomegaly. The finding of the mutation, described as c.804_805insTA in sequence NM_001173517.1, hemizygous and inherited from the heterozygous mother, led to the authors to propose to MAP7D3 as a candidate gene for the patient disorder considering the nature of the novel and truncating variant identified as well as the probable involvement of MAP7D3 in neurodevelopmental disorder, a speculation supported by the implication of MAP7 protein in neuronal morphogenesis [5], role of MTs in regulating neurons and therefore brain development [6] and growing evidence of association of mutations in MT-associated proteins to different neurodevelopmental disorders, including intellectual disability and ASD [6-10]. In this way, an involvement in intellectual disability has been reported for kinesin genes, motor proteins that control both intracellular cargos transport and MTs cytoskeleton organization. Among them, for KIF4A and KIF5C [11]. In case of KIF4A of Xq13.1 observed in 4 affected males of a family with X-linked mild to moderate mental retardation, poor speech and seizures (described in OMIM as Intellectual developmental disorder, X-linked 100; https://www.omim.org/entry/300923) all carriers of hemizygous KIF4A mutation c.1489-8_1490delins10 (in sequence NM_012310.4) resulting in disruption of the exon 15 acceptor splice site leading to an in-frame deletion of exon 15 from the protein, a genotype that segregated with the disorder in the family [11]. Besides, KIF5B coding for kinesin-1 interacting with that of MAP7D3 has been proposed as a candidate gene for neurological diseases following the identification of homozygous KIF5B missense variant p.His751Arg (NM_004521.3:c.2252A>G) that segregated in two brothers with a disorder including developmental and speech delay [12].
Our data would reinforce the concept of a neurodevelopmental disorder due to haploinsufficiency of MAP7D3, according to X-Linked Recessive (XLR) inheritance. It would constitute a disorder associated with a specific function of MAP7D3, excluding redundant functions of MAP7 proteins, whose precise delineation requires necessarily more data from replication studies and gene functions. It would probably be compatible with the continuum of a moderate neurodevelopmental phenotype without congenital cerebral malformations. In this regard, the lack of Autism (ASD) in the Saudi Arabia patient, but present in our patient, may represent genotype-phenotype association differences attributable to the location of mutations and the predicted protein alteration. Nonsense mutation D304X of Saudi Arabia patient would yield truncated short molecules presumably fully defective, at least for interactions with kinesin-1. By contrast, frameshift mutation T821RfsX8 of our patient would yield larger and partially functional MAP7D3 molecules, although defective in the MDCT domain, retaining the MAP or kinesin-1 binding region (MD) but lacking the C-Terminal Tail (CT) that according to results of in vitro assays would have a role in regulating microtubule assembly and stability [1-3].
Ethical Approval
Authors get parental permission for publication of patient and family data.
Conflicts of Interest
They declare that there is no conflict of interests regarding the publication of this paper.
References
- Sun X, Shi X, Liu M, Li D, Zhang L, Liu X, Zhou J. Mdp3 is a novel microtubule-binding protein that regulates microtubule assembly and stability. Cell Cycle. 2011 Nov 15;10(22):3929-37. doi: 10.4161/cc.10.22.18106. Epub 2011 Nov 15. PMID: 22142902.
- Yadav S, Verma PJ, Panda D. C-terminal region of MAP7 domain containing protein 3 (MAP7D3) promotes microtubule polymerization by binding at the C-terminal tail of tubulin. PLoS One. 2014 Jun 13;9(6):e99539. doi: 10.1371/journal.pone.0099539. PMID: 24927501; PMCID: PMC4057234.
- Hooikaas PJ, Martin M, Mühlethaler T, Kuijntjes GJ, Peeters CAE, Katrukha EA, Ferrari L, Stucchi R, Verhagen DGF, van Riel WE, Grigoriev I, Altelaar AFM, Hoogenraad CC, Rüdiger SGD, Steinmetz MO, Kapitein LC, Akhmanova A. MAP7 family proteins regulate kinesin-1 recruitment and activation. J Cell Biol. 2019 Apr 1;218(4):1298-1318. doi: 10.1083/jcb.201808065. Epub 2019 Feb 15. PMID: 30770434; PMCID: PMC6446838.
- Monies D, Abouelhoda M, AlSayed M, Alhassnan Z, Alotaibi M, Kayyali H, Al-Owain M, Shah A, Rahbeeni Z, Al-Muhaizea MA, Alzaidan HI, Cupler E, Bohlega S, Faqeih E, Faden M, Alyounes B, Jaroudi D, Goljan E, Elbardisy H, Akilan A, Albar R, Aldhalaan H, Gulab S, Chedrawi A, Al Saud BK, Kurdi W, Makhseed N, Alqasim T, El Khashab HY, Al-Mousa H, Alhashem A, Kanaan I, Algoufi T, Alsaleem K, Basha TA, Al-Murshedi F, Khan S, Al-Kindy A, Alnemer M, Al-Hajjar S, Alyamani S, Aldhekri H, Al-Mehaidib A, Arnaout R, Dabbagh O, Shagrani M, Broering D, Tulbah M, Alqassmi A, Almugbel M, AlQuaiz M, Alsaman A, Al-Thihli K, Sulaiman RA, Al-Dekhail W, Alsaegh A, Bashiri FA, Qari A, Alhomadi S, Alkuraya H, Alsebayel M, Hamad MH, Szonyi L, Abaalkhail F, Al-Mayouf SM, Almojalli H, Alqadi KS, Elsiesy H, Shuaib TM, Seidahmed MZ, Abosoudah I, Akleh H, AlGhonaium A, Alkharfy TM, Al Mutairi F, Eyaid W, Alshanbary A, Sheikh FR, Alsohaibani FI, Alsonbul A, Al Tala S, Balkhy S, Bassiouni R, Alenizi AS, Hussein MH, Hassan S, Khalil M, Tabarki B, Alshahwan S, Oshi A, Sabr Y, Alsaadoun S, Salih MA, Mohamed S, Sultana H, Tamim A, El-Haj M, Alshahrani S, Bubshait DK, Alfadhel M, Faquih T, El-Kalioby M, Subhani S, Shah Z, Moghrabi N, Meyer BF, Alkuraya FS. The landscape of genetic diseases in Saudi Arabia based on the first 1000 diagnostic panels and exomes. Hum Genet. 2017 Aug;136(8):921-939. doi: 10.1007/s00439-017-1821-8. Epub 2017 Jun 9. PMID: 28600779; PMCID: PMC5502059.
- Tymanskyj SR, Yang BH, Verhey KJ, Ma L. MAP7 regulates axon morphogenesis by recruiting kinesin-1 to microtubules and modulating organelle transport. Elife. 2018 Aug 22;7:e36374. doi: 10.7554/eLife.36374. PMID: 30132755; PMCID: PMC6133550.
- Matamoros AJ, Baas PW. Microtubules in health and degenerative disease of the nervous system. Brain Res Bull. 2016 Sep;126(Pt 3):217-225. doi: 10.1016/j.brainresbull.2016.06.016. Epub 2016 Jun 27. PMID: 27365230; PMCID: PMC5079814.
- Bonini SA, Mastinu A, Ferrari-Toninelli G, Memo M. Potential Role of Microtubule Stabilizing Agents in Neurodevelopmental Disorders. Int J Mol Sci. 2017 Jul 26;18(8):1627. doi: 10.3390/ijms18081627. PMID: 28933765; PMCID: PMC5578018.
- Ramkumar A, Jong BY, Ori-McKenney KM. ReMAPping the microtubule landscape: How phosphorylation dictates the activities of microtubule-associated proteins. Dev Dyn. 2018 Jan;247(1):138-155. doi: 10.1002/dvdy.24599. Epub 2017 Oct 27. PMID: 28980356; PMCID: PMC5739964.
- Lasser M, Tiber J, Lowery LA. The Role of the Microtubule Cytoskeleton in Neurodevelopmental Disorders. Front Cell Neurosci. 2018 Jun 14;12:165. doi: 10.3389/fncel.2018.00165. PMID: 29962938; PMCID: PMC6010848.
- Kalantari S, Filges I. 'Kinesinopathies': emerging role of the kinesin family member genes in birth defects. J Med Genet. 2020 Dec;57(12):797-807. doi: 10.1136/jmedgenet-2019-106769. Epub 2020 May 19. PMID: 32430361; PMCID: PMC7691813.
- Willemsen MH, Ba W, Wissink-Lindhout WM, de Brouwer AP, Haas SA, Bienek M, Hu H, Vissers LE, van Bokhoven H, Kalscheuer V, Nadif Kasri N, Kleefstra T. Involvement of the kinesin family members KIF4A and KIF5C in intellectual disability and synaptic function. J Med Genet. 2014 Jul;51(7):487-94. doi: 10.1136/jmedgenet-2013-102182. Epub 2014 May 8. PMID: 24812067.
- Charng WL, Karaca E, Coban Akdemir Z, Gambin T, Atik MM, Gu S, Posey JE, Jhangiani SN, Muzny DM, Doddapaneni H, Hu J, Boerwinkle E, Gibbs RA, Rosenfeld JA, Cui H, Xia F, Manickam K, Yang Y, Faqeih EA, Al Asmari A, Saleh MA, El-Hattab AW, Lupski JR. Exome sequencing in mostly consanguineous Arab families with neurologic disease provides a high potential molecular diagnosis rate. BMC Med Genomics. 2016 Jul 19;9(1):42. doi: 10.1186/s12920-016-0208-3. PMID: 27435318; PMCID: PMC4950750.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.