Biology Group . 2022 August 09;3(8):867-879. doi: 10.37871/jbres1525.
Mechanisms of PD-L1 Regulation in Tumor Immune Ecosystem
Fatima Bangash*, Sabiha Alia and Rabia Gul
- Programmed death receptor-1
- Programmed death-ligand 1
- Gene expression regulation
- Immune checkpoints blocking therapy
Abstract
PD-1 (Programmed Death Receptor-1) is a cell membrane protein found on Cytotoxic T cells (CTLs) surface. An abundantly expressed type I transmembrane protein in healthy tissues, Programmed Death-Ligand 1 (PD-L1), has a molecular mass of 40 kDa. The extracellular binding of the proteins PD-1 and PD-L1 prevents the development of autoimmune disorders by suppressing the activation of CTLs under normal physiological conditions. It has been shown that cancer cells can avoid immune monitoring by increasing PD-1/PD-1-mediated CTL inactivation in melanoma, lung cancer, renal cell carcinoma, and other malignant tumors. PD-L1 expression regulation has been described in recent years from the standpoint of gene amplification, chromatin modification, post-transcription and transcription modification, translation, and post-translational modification. Anti-PD-1 immunotherapy has demonstrated promising results in treating several cancers, including breast, lung, and prostate cancers. This review aims to present the most recent research findings in PD-L1 regulation in cancer cells. A tumor immunotherapy strategy targeting PD-1 and PD-L1 is expected to be useful.
Introduction
The structural backbone of PD-L1
Regarding immunoglobulins, one of the most common members is PD-L1(Programmed death-ligand 1), also known as B7-h1 or Cluster of Differentiation 274 (CD274). An extracellular portion, a hydrophobic transmembrane sequence, and an intracellular area make up this type I transmembrane protein, which belongs to the family of transmembrane proteins. The extracellular domain matrix domain of PD-L1 contains two immunoglobulin structures: the IgV distal area and the IgC proximal region. The IgV sequences have a typical Ig-like domain with a CDR that allows PD-L1 to bind to PD-1 1:1 [1]. Pd-l1 is attached to the cell membrane by hydrophobic transmembrane regions, and its intracellular structure is unique among B7 family members. These intracellular motifs are RMLDVEKC, DTSSK, and QFEET. These three motifs are relatively conserved in mammalian PD-L1 molecules, and signal transmission is likely mediated by RMLDVEKC and DTSSK motifs. For example, the intracellular region of PD-L1 protein in mouse cells contains two lysine residues. For example, it is possible to control how stable and effective PD-signal L1's transduction is by changing lysine residues on the protein [1]. In addition to CD4+ and CD8+ T cells, Pd-1 is found on macrophages, dendritic cells, and other cells. Immunoreceptor Inhibitory Motif (ITIM) is found at the N-terminus of the intracellular domain, which includes an IgV region. C-terminal tyrosine-based Immunoreceptor Switch Motif (ITSM) was designed.
PD-L1 regulation in tumor immune cells
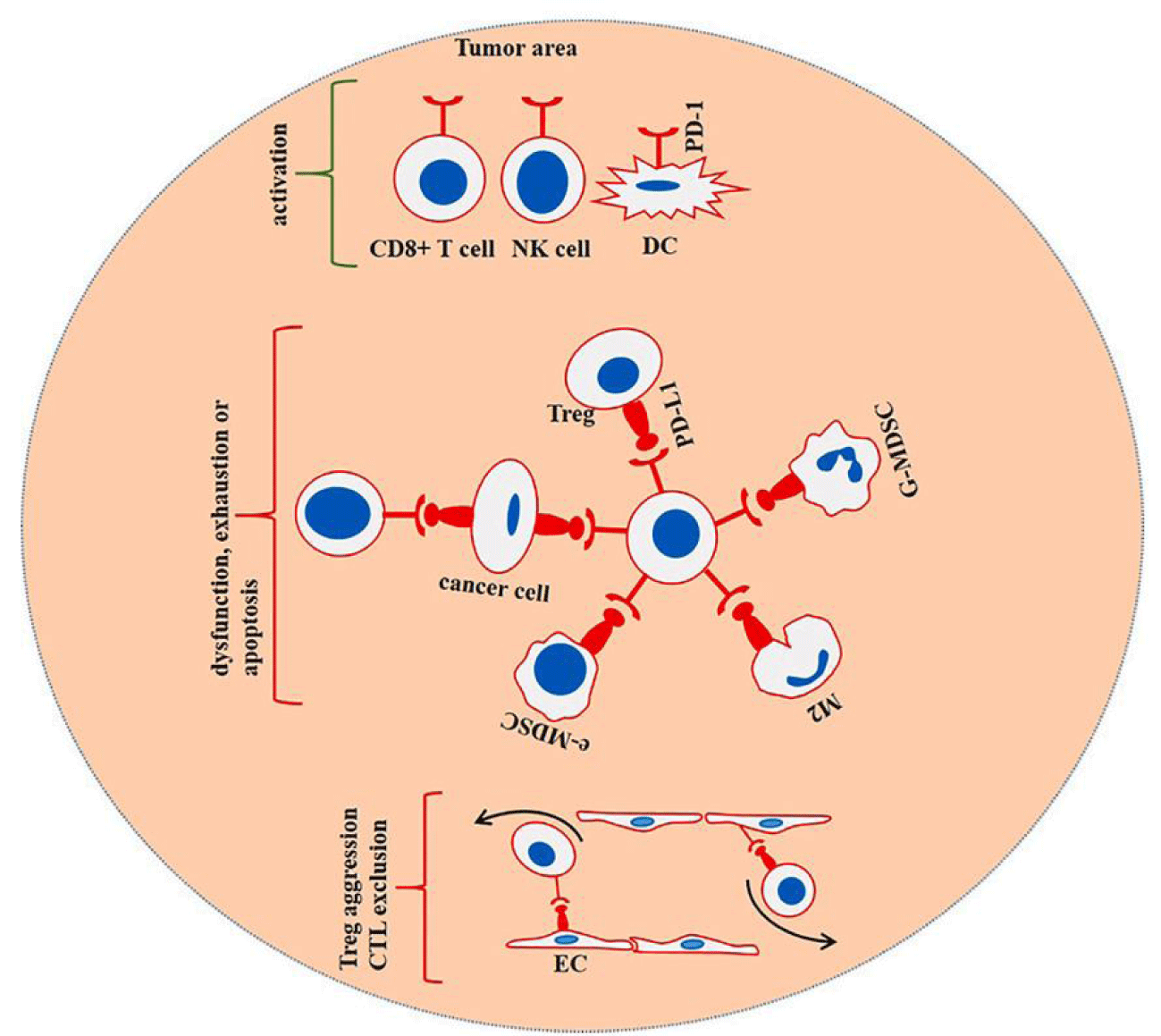
T cells are the primary effector cells in the body's immune response after being acquired. Foreign antigens collected in lymph nodes or the spleen trigger an immunological response under normal circumstances. The Dendritic Cell (DC) is one type of myeloid cell that can be thought of as a cell that presents antigens. When the antigen-specific T Cell Receptor (TCR) on the surface of the T cell recognizes and binds to the antigenic peptides produced by the Antigen-Processing Centre (APC), an immune response to the antigen has elicited the proliferation of antigen-specific T cells. T cells require "co-stimulation" to become activated, in addition to the binding of the TCR to peptide and MHC. T cell receptors CD80(b7-1) and CD86(b7-2) attach to APC proteins of the B7 family-like CD80(b7-1) and CD86(b7-2), which bind to CD28 on T cells.
Furthermore, give "co-stimulatory" signals that stimulate T cells upon antigen detection of immunological synapses [1]. These signals can rescue T cells from apoptotic signals provided by PD-1 and PD-L1 while stimulating the TCR to deliver proliferative signals. Interleukin-10 secretion is triggered when PD-1 and PD-L1 attach to the surface of T Lymphocytes (IL-10), while the secretion of Interleukin-2 (IL-2) is caused by abnormal B7 family proteins [2]. It was proved that PD-1/PD-L1 could conduct inhibitory signals through IL-10, reduce the proliferation of T cells in lymph nodes and induce the production of apoptotic signals. Under the normal mechanism, PD-1/PD-L1 can prevent oversensitivity caused by the excessive immune response, but it is related to the immune escape mechanism of tumor cells. Many in vivo tumors and cancer cell lines reverse induce effector T cell apoptosis by expressing PD-L1, resulting in a strong suppression of the immune response of T cells in human neoplastic diseases, and their exosomes are highly correlated with the proliferation of tumor cells. Therefore, PD-L1 is widely regarded as an important immune checkpoint for cancer progression and poor prognosis.
The regulation mechanism of PD-L1 expression is multi-dimensional. This paper reviews the regulation mechanism of DNA induction, transcription, translation, post-translational modification, and cell microenvironment and discusses the regulation of PD-L1 expression by introducing small-molecule drugs.
Regulation mechanism of PD-L1 expression at DNA level
Chromosome 9P24.1 encodes the gene for PD-L1. Acetylation of histones in this region triggers PD-L1 expression. When HDAC inhibitors are used, they increase histone acetylation at roughly 455 base pairs upstream of the first PD-L1 gene exon and relax chromatin [3,4]. This increases the PD-L1 gene's expression. In addition, methylated DNA can recruit HDAC, thus inhibiting PD-L1 gene transcription and down-regulating PD-L1 expression. Therefore, some DNA methyltransferase inhibitors also potentially enhance PD-L1 expression. As with HDAC inhibitors, azacitidine and decitabine have been shown to improve PD-L1 expression in human melanoma cell lines and mice models of the disease [3]. Other studies have shown that HDAC6 can signal and activate transcription 3. STAT3 enforces the PD-L1 promoter and activates the stat3 signal to enhance the expression of PD-L1. In the absence or inhibition of HDAC6, protein phosphatase 2A(PP2A) interacts with STAT3 more. In turn, pp2A-mediated dephosphorylation of STAT3 was promoted, and the phosphorylation level of STAT3 decreased [4]. When Interleukin-6 (IL-6) stimulated human melanoma cells, the increase of PD-L1 expression in hdAC6-knockout cells was significantly reduced. Therefore, the STAT3 signalling pathway may be an important factor affecting PD-L1 expression [4]. In addition, Bromodomain and Extra Terminal Domain (BET) family proteins are histone acetylation readers that can promote PD-L1 gene transcription by directly binding to acetylated lysine in histone tails. A member of the BET family, Bromodomain Contained Protein 4 (BRD4), has been identified as a target of CD274, which can directly mediate transcription of the CD274 gene. It has been shown that BRD4 can efficiently boost PD-L1 expression by binding to acetylated histone H3K27Ac in the PD-L1 promoter region and the distal enhancer of the gene. In ovarian cancer models, because the BRD4 gene is situated on chromosome 19P13.1, and this region is frequently amplified in this malignancy, suppression of BRD4 protein can greatly limit the production of PD-L1 and expression and activity of cytotoxic T cells. According to animal studies, the BET inhibitor JQ1 dramatically decreased PD-L1 expression in tumor cells, tumor-associated dendritic cells, and macrophages, while the BRD4 inhibitor has been demonstrated to suppress the interferon-induced elevation of PD-L1 expression. To summarize, it appears that BRD4 is a critical player in regulating PD-L1 expression and that its presence may influence how well PD-L1 responds to signals from the Tumor Microenvironment (TME) [5].
Gene mutation is an important factor affecting protein expression and function. For example, some mutations may lead to protein folding disorder, damaging the interaction between PD-1 and PD-L1 and inhibiting the immune escape pathway. It may also increase the affinity of the two proteins, thereby enhancing or weakening the effect. Studies in molecular structure have shown that the N-terminal of PD-L1 has a V-folding domain similar to Ig, responsible for its binding with PD-1 [6]. The Ig-like V-domain of Pd-l1 binds with PD-1 via a broad hydrophobic surface. It is possible to combine the "front end" of PD-1 with PD-L1 by aligning the long axes of PD-1 and PD-L1 approximately perpendicularly in the complex [6]. There are two ways PD-L1 (primarily Tyr123) and its nearby residues drive PD-1 side chains toward the protein's core, which results in fissures in its contact surface with Tyr123 and the surrounding residues. This induced fitting adds to the number of interaction surfaces available. The likelihood of PD-1 and PD-L1 interacting was improved [6]. There have been two reports of PD-1 mutations. Because of the differences in ring configurations between the BC, CCC, and FG chains in the high-affinity PD-1 mutants, the contact force between PD-1 and the Antigen-Presenting Cell (APC) is greatly increased. Three specific mutations have the potential to stabilize the PD-1-PD-L1 interaction: M70E (Y68H), K78T (K78T), and M70E (M70E). PD-1 mutates penetrate larger tumors better than PD-L1 monoclonal antibodies, making them a more effective immune system modulator [7]. Another PD-1 mutant was created by substituting only one amino acid (A132L), and the binding of the PD-1 mutant to PD-L1 was found to be 45 times stronger than that of the wild-type PD-1 [8]. This increased the van der Waals force required for binding the mutant to PD-L1 and the mutant's affinity to PD-L1 by 45 times [8]. PD-crystal L1's structure has also been studied, and the results suggest that the protein contains small molecule inhibitor sites. In the case of small-molecule PD-1/PD-L1 inhibitors, such as those developed by BMS (Bristol Myers Squibb), these molecules can bind to the surface where PD-1 interacts with PD-L1 as a result of the dimerization of PD-L1. Antibody checkpoints PD-1/PD-L1 could benefit from adding small-molecule medicines that can connect between albumin molecules while creating protein dimers. Examples include the small-molecule inhibitors, BMS-202 and BMS-8, which bind to PDl1, preventing the interaction of PD-1 with PDl1 [6,9]. G to C substitution at SNP 4143815, which is prevalent in gastric cancer, can increase PD-L1 expression and enhance the ability of cancer cells to survive and invade by disrupting mir–570–mediated post-recording/translation regulation [10]. Another thing that changes the expression of PD-L1 is when other related functional genes are deleted. For example, jak1/2 functional deletion mutations do not make PD-L1 reactive and do not change the genetic mechanism of how interferon - works, which makes PD-L1 resistant to PD-L1 blocking treatment, as mentioned in figure 1.
Many proto-oncogenes upregulate their protein expression by changing copy numbers. According to a study published in the Journal of Clinical Pathology, patients with small-cell lung cancer who have CD274 local amplification are more likely to express PD-L1 at high levels than those who do not. An increase in CD274 transcripts and an increase in the expression of PD-L1 are induced by a genomic rearrangement [11,12]. Chromosome 9P24 amplification has increased PD-L1 expression. NSCLC (Non-Small Cell Lung Cancer) has also been associated with gene 1 [13]. Genome 9P24 was related to an increased expression of Janus Kinase 2. Although the PD-L1 protein expression level was still elevated when JAK2 inhibitor TG-101348 and STAT3 inhibitor BP-1-102 were used, they greatly reduced the PD-L1 copy number amplification and up-regulation. However, PD-L1 protein expression was not affected by the STAT3 inhibitor fludarabine [14]. This shows that the JAK2 and PD-L1 genes may be trans-activated.
On the other hand, proto-oncogenic MYC ordinarily encodes a transcription factor that either stimulates or suppresses the expression of its target gene, but under certain circumstances, it can be activated ontogenetically through amplification or translocation. Studies have shown that the overexpression of MYC is a characteristic of many human cancers, and the overexpression of MYC will directly lead to the malignant transformation of various cell types. In addition to driving the occurrence and development of tumors, MYC also plays a role in tumor maintenance. In several myC-driven tumor mouse models, the survival of tumor cells depends on continuous MYC overexpression, and restricting MYC expression or blocking its function can lead to growth stagnation, apoptosis, or differentiation of tumor cells [15]. The MYC gene appears to function in PD-L1 transcription in two ways. MYC binds to the promoter of the PD-L1 gene in teT-off MYC transgenic mice models of acute T Lymphoblastic Leukemia (T-all) or Hepatocellular Carcinoma (HCC) cell types, increasing PD-L1 expression. Similar findings were made in human T-ALL cell lines, HCC cell lines HepG2, melanoma cell lines SKMEL28, and H1299, including in T-ALL primary tissues [16].
On the other hand, MYC did not affect both breast cancer cells AT3 and lymphoma cells E-MyC. Even in the TetOFF mouse liver cancer model, MYC can decrease PD-L1 production regardless of whether the Interferon-Alpha (IFN-) is exposed [16]. Researchers have not yet reached an accurate conclusion on the systemic regulatory role of MYC, and its complex regulatory mechanism is still the focus of future research. In conclusion, gene amplification is one of the factors driving PD-L1 expression in tumors, but it is not the decisive factor.
Structural Variations (SVs), including translocation, inversion, tandem replication, and deletions, have been widely observed in cancer genes and are an important factor affecting the alteration of protein expression. The 3 '-Untranslated Region (3' -UTR) of PD-L1 transcription negatively regulates the stability of mRNA. SVs usually destroy the 3 '-terminal domain of the PD-L1 gene, resulting in abnormal activation of PD-L1 and immune escape. These transcribed mRNA can be stabilized by truncating 3 '-UTR. The underlying mechanism may be that some cis-acting elements in mRNA decay in the 3' -UTR region are damaged, including Au-rich elements and potential miRNA(microRNA) attachment sites. The findings revealed that truncation of the 3'-UTR of PDL1 was associated with enhanced PD-L1 expression in a statistically significant and independent manner. It was also found that PD-L1 expression was upregulated more by 3'-UTR truncation than by interferon stimulation. It has been shown that cells with damaged 3' -UTRs may more effectively regulate PD-L1 expression when IFN- and PD-L1 3 '-UTR truncation act synergistically [17], highlighting the critical role of 3 '-UTR in the expression of PD-L1. SV(+) samples had a significantly lower antitumor immune response than SV(-) samples with similar PD-L1 levels, indicating a decreased ability of effector T cells to destroy tumor cells. Meanwhile, when contrasted to wild-type PD-L1 mRNA, PD-L1 expression was significantly increased after 3 '-UTR truncation, and mRNA clearance was also significantly delayed, indicating that PD-L1 3' -UTR also plays an important role in mRNA stability [17]. Cancers caused by a viral infection can also cause abnormal transcription of PD-L1 by integrating genes. For example, in cervical squamous cell carcinoma, Human Papillomavirus (HPV) integrates into CD274, extending transcription from truncated PD-L1 to HPV E2 and E5 genes. Significantly amplified CD274 allele disrupted by the virus [17]. It has also been discovered that Epstein-Barr virus-transformed lymphoblastic cell lines exhibit increased production of the PD-L1 protein, which is regulated via the LATENT Membrane Protein 1 (LMP1), JAK/STAT-dependent promoter and activator protein-1, and LATENT Membrane Protein 1 (LMP1) (LMP1). Enhancer activity linked to Ap-1) [18]. EBV latent membrane protein 1 has been shown to promote PD-L1 expression via the LMP1/AP-1/Nuclear factor kappab (NF- B) pathway and synergize with IFN- [19]. Viral integration in virus-mediated malignancies may cause evasion of antiviral immunity at the onset of infection and subsequent immune surveillance by overexpression of PD-L1 in infected cells [17]. In addition, NF-κB inhibitors effectively attenuated PD-L1 induction, suggesting that NF-κB is an essential signalling pathway for lMP1-induced PD-L1 expression [9]. Therefore, NF-κB is also an important site blocked in cancer therapy.
Regulation mechanism of PD-L1 expression in transcription and post-transcription
Pd-l1 can initiate transcription after activating multiple signalling pathways, such as hypoxia-inducible factor-α (HYPOxia inducible factor-α), which has been reported. HIF-, proto-oncogene MYC, STATs, NF-B, and AP-1 can bind to PD-L1-related genes and promote translation of the PD-L1 protein. Transcription factors such as IFN-/JAKs / STAT1 / Interferon Regulatory Factor (IRF) and other signalling pathways, as well as lactic acid-rich microenvironment regulation [9], are transduced via these signalling pathways. A slew of transcription factors implicated in the control of PD-L1 expression was discovered. Firstly, the PD-L1 gene enhancer region promotes transcriptional activity by binding to the AP-1 component of the transcription factor AP-1 in typical CHL Reed-Sternberg cells. This increases the activation of the PD-L1 gene [18]. Secondly, studies have shown that the transcription factor expressed by the IRF1 gene is necessary to regulate the expression of PD-L1 in interferon-γ-induced cancer cells, and the combination of IRF1 and its promoter is conducive to the maintenance of interferon-γ-induced effect and can enhance interferon-induced PD-L1 expression [20].
The transcription factor NF-κB is also required to produce PD-L1 in cancer cells. Prostate and colon cancer Tumor Necrosis Factor (TNF) stimulates the production of PD-L1 (PD-L1) through the NF-B signalling pathway [16,21,22]. It was discovered that the PD-L1 gene's promoter region and its 140 Kb downstream enhancer region have NF-B binding sites. It is also crucial to note that STAT3 is another transcription factor that, by binding to the PD-L1 promoter, increases the expression of PD-L1. STAT3 silencing has been proven to be effective in removing oncogenic chimeric nucleophosmin. The elevation of PD-L1 protein expression is caused by the NPM/ANAplastic Lymphoma Kinase (ALK) mutation [23], indicating that STAT3 may play a role in immunological evasion. The IFN-/JAKs/STATs/IRF1 axis, type II interferon, has been demonstrated to be the major regulator of PD-L1 expression in studies. In numerous forms of cancer, IFN- is the principal inducer of PD-L1, IFN- via the JAK/STAT1/IRF1 pathway. Melanoma, non-small cell lung cancer, liver cancer, squamous cell carcinoma of the head and neck, gastric cancer, myeloma, and glioma are just a few examples [16,20,24-28]. The phosphorylation of JAK1 and JAK2 occurs when IFN- binds to the receptor. In most cells, phosphorylation of the receptor causes STAT1 to attach and phosphorylate to form a dimer, but in some cells, phosphorylation of STAT3 causes STAT1 to attach and phosphorylate to form a dimer. Pd-l1 expression was considerably boosted by binding and activating related genes. The activated dimers aggregate in the nucleus and act as transcription factors that bind to IFN-induced genes with Gamma Interferon Activation Sites (GAS), such as the IRF1 gene. Some negative regulators of interferon, such as Suppressors of Cytokine Signalling (SOCS), bind to JAK2 to signal and participate in the negative feedback regulation of cytokines. Thus, decreased STAT1 and STAT3 activities affect IFN-γ induction [20].
Inflammatory cytokine Epidermal Augmented Factor (EGF) is a 53 amino acid polypeptide that is heat resistant and has low molecular weight. It stimulates the production of epidermal growth factors by phosphorylating the enzyme phosphatidylinositol-3-kinase. Head and neck squamous cell carcinoma and Non-Small-Cell Lung Cancer (NSCLC) tumors express PD-L1 in response to the PI3K/Protein kinase B (PKB/AKT)/Mammalian target of rapamycin mTOR and JAK2/STAT1 pathways, as well as epidermal augmented factor receptor, activation mutation of EGFR gene, and mutation of Echinoderm Microtubule-Associated Protein-like 4 (EML4) gene [16,26,29-31]. Meanwhile, GSK3β, the isomer of GSK3α, promotes the degradation of PD-L1 by phosphorylation of two phosphorylation sites, T180 and S184, in the extracellular area of PD-L1. Therefore, GSK3α and GSK3β contribute to antitumor immune escape [32-34].
In addition to the above induction factors, many other substances significantly regulate the activation of PD-L1 in specific cells. Toll-like receptor 3(TLR3) can induce the up-regulation of PD-L1 expression in 2426 · Review · neurocytoma [35]; The ARE binding protein TTP(Tristetraprolin) can negatively regulate the expression of PD-L1 through the 3 '-UTR of PD-L1 mRNA rich in AU elements, and RAS signal can also lead to TTP phosphorylation through MEK signal downstream. PD-L1 expression in cancer cells can be boosted by inhibiting TTP via the mitogen-activated protein kinase 2 (MK2) pathways [36]. Ubiquitin C-terminal hydrolase L1(UCHL1) is an abnormally expressed regulator of cell signal transduction in NSCLC. UCHL1 can also promote the expression of PD-L1 in NSCLC cell lines by activating the AKT/P65 signalling pathway, suggesting that the inhibition of UCHL1 may inhibit the immune escape of NSCLC by down-regulating the expression of PD-L1 in NSCLC cells [37]. Endogenous transcriptional regulatory factor nucleophosmin 1(NPM1) binds to the PD-L1 promoter to enhance its activity in triple-negative breast cancer (TNBC) cells.
Moreover, it increases the mRNA and protein expression of PD-L1 [38]. Studies on the modulation of PD-L1 by specific substances in specific cells are numerous and not systematic. More in-depth studies are still needed to have a prospect of development and application.
Pd-l1 translation and the regulatory mechanism of post-translational modification
MiRNAs are a class of highly conformed small non-coding RNAs that regulate gene expression at the post-transcriptional level and potentially regulate various aspects of cell activity, including differentiation and development, metabolism, proliferation, apoptosis, viral infection, and tumorigenesis. Mir-513 was shown to regulate PD-L1 translation and IFN-γ-induced expression of PD-L1 in human bile duct cells. Pd-l1 mRNA was expressed, but PD-L1 protein was not expressed in resting human bile duct cells. IFN-γ can induce the expression of PD-L1 protein and change the expression profile of miRNA in bile duct cells. Among miRNAs with IFN-γ down-regulation function, Mir-513 is complementary to the 3 '-UTR of PD-L1, and targeting Mir-513 can inhibit the translation of PD-L1 mRNA. However, PDL1 mRNA was not degraded [39,40]. The complementary sequence of Mir-34a also exists in the 3 '-UTR of PD-L1 mRNA. Studies have shown that Mir-34A is involved in inhibiting the expression of IFN-γ and As2O3 induced human leukemia cell PD-L1 and inducing T cell apoptosis. Moreover, it was confirmed that Mir-34a significantly inhibited the expression of PD-L1 in HL-60 cells [41], and the above evidence proved that Mir-34a could also target the expression of PD-L1. At the same time, Mir-34A is regulated by P53, which participates in immune responses by regulating inflammatory cytokines, toll-like receptors and interferon signal transduction, and activation of T cells and NK cells. P53 can directly bind to the 3 '-UTR of PD-L1 through Mir-34 and specifically regulate tumor immune response by regulating the expression of PD-L1, and Mir-200, which can directly regulate the expression of PD-L1, has also been proved to be regulated by P53 [42]. Mir-200 is an epithelial-to-mesenchymal transition (EMT) inhibitor targeting PD-L1. Zip-finger ebox-binding homeobox 1(Zip-finger ebox-binding homeobox 1) is an EMT activator and can weaken the inhibition of Mir-200. Studies have shown that Mir-200 /ZEB1 is an EMT regulatory axis. Pd-l1 is the downstream target of the Mir-200 /ZEB1 axis, so the expression of PD-L1 in tumor cells can be regulated by regulating the mir-200 /ZEB1 axis [43]. A MIR-197 /CDC28 protein kinase regulatory subunit 1B(CKS1B)/ STAT3-mediated PD-L1 network has been reported in chemotherapy-resistant NSCLC. This network is independent of immunosuppressive signals, and the expression level of Mir-197 is negatively correlated with the expression of PD-L1. With the down-regulation of Mir-197, drug resistance and metastasis of NSCLC cells in vivo and in vitro are enhanced. Mechanism studies suggest that the Mir-197-mediated CKS1B/STAT3 axis plays a role in tumor progression regulated by multiple genes (Bcl-2, C-MyC, and Cyclin D1), PD-L1 may be a biomarker of this axis. It was suggested that Mir-197 could regulate drug resistance and tumor progression of NSCLC through CKS1B and STAT3, proving that Mir-197 /CKS1B/ STAT3-mediated signalling drives tumor PD-L1 expression [44]. Similarly, it was found in Lung Adenocarcinoma (LUAD) that Mir-326 inhibited the gene expression of LUAD immune checkpoint molecules PD-L1 and B7-H3, changed the invasion degree of CD8+T cells, and reduced the migration ability of tumor cells [45]. Mir-424 (322)[46], Mir-570 [10], Mir-138-5p [47], Mir-17-5p [48], Mir-25-93-106b [49], etc., have also been reported, but they are independent studies for different cancer types. Whether they have a synergistic effect on these miRNAs is uncertain.
In addition to regulating PD-L1 translation through miRNA, mTOR activation also regulates PD-L1 protein synthesis in cancer cells. The PD-l1 expression depends on mTOR in glioma, breast, prostate, ovarian and pancreatic cancers studies. Studies have shown that the activation of the AKT/mTOR pathway strictly regulates the expression of PD-L1 in vitro and in vivo. The activation of mTOR increases the expression level of PD-L1 protein but does not affect the mRNA level of PD-L1. Therefore, regulation of PD-L1 by this pathway is considered to occur at the level of translation [50]. Loss of PTEN in glioma cells leads to activation of THE PI3K/AKT/mTOR/S6K1 pathway, resulting in increased PD-L1 protein level. As mentioned above, the PTEN product is an important tumor suppressor and inhibitor of the PI3K/AKT signal transduction pathway, which can inhibit the expression of PD-L1 in breast adenocarcinoma cells. In addition, studies have shown that constitutive expression of PD-L1 is also affected by mutations of oncogenes EGFR, KRAS, and ALK [50].
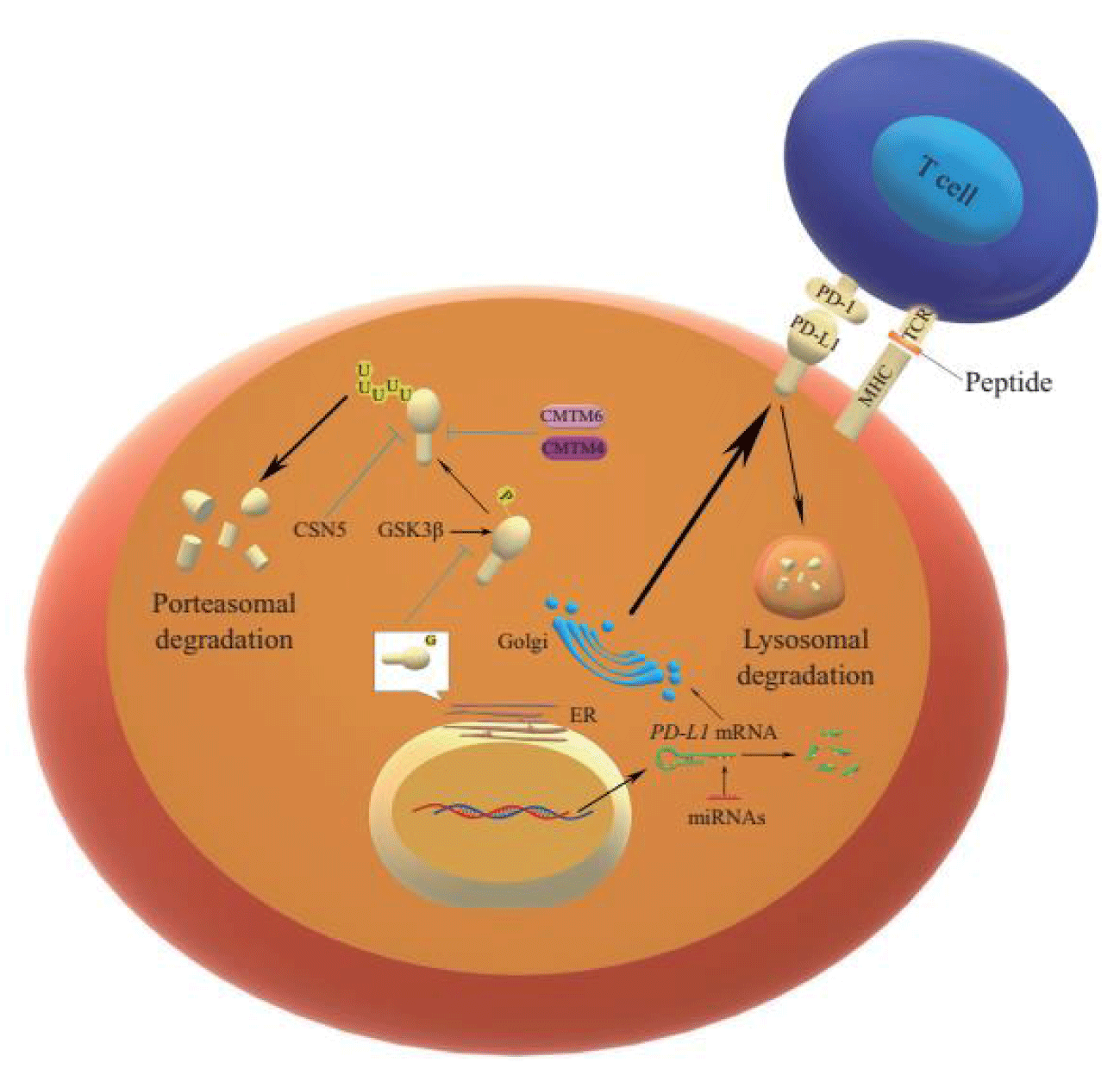
In addition to regulating PD-L1 translation, post-translational modifications such as glycosylation, phosphorylation, ubiquitination, and deubiquitination of PD-L1 protein and PD-L1 binding proteins and protein subcellular transport processes can also affect the stable expression of PD-L1 protein in tumor cells [16]. N- glycosylation is an important modification of the white matter that determines protein structure and function. By changing protein conformation, glycosylation can regulate protein activity and protein-protein interaction. Pd-l1, as a membrane protein, is extensively glycosylated after translation. N-glycosylation of the extracellular region of PD-L1 occurs in the lumen of the Endoplasmic Reticulum (ER), and this modification promotes the interaction between PD-L1 and lipid membranes. In Western blot analysis, glycosylated PD-L1 was detected at 45 kDa, while non-glycosylated PD-L1 was detected at 33 kDa. Through bioinformatics prediction, mass spectrometry, and mutagenesis, it was found that N-glycosylation of PD-L1 occurred only at N35, N192, N200, and N219 [51]. Glycogen Synthase kinase 3β(GSK3β), a glycogen synthase kinase 3β(GSK3β), forms a glycogen synthase kinase 3β(GSK3β) binding region containing N192, N200, and N219 residues, which are masked by n-glycosylation. They destroy the interaction between PD-L1 and GSK3β and inhibit the phosphorylation of PD-L1 caused by GSK3β and the subsequent β-transduction repeat-containing proteins. β-TrCP) mediated ubiquitination (Figure 1) [51]. Glycosylated PD-L1 cannot interact with GSK3β, thus affecting the phosphorylation process of PD-L1, so the degree of glycosylation of PD-L1 protein is affected. Meanwhile, glycosylation also indirectly prevented the ubiquitination and degradation of the PD-L1 protein. Most PD-L1 in cancer cells is glycosylated and cannot be degraded by proteasomes [16].
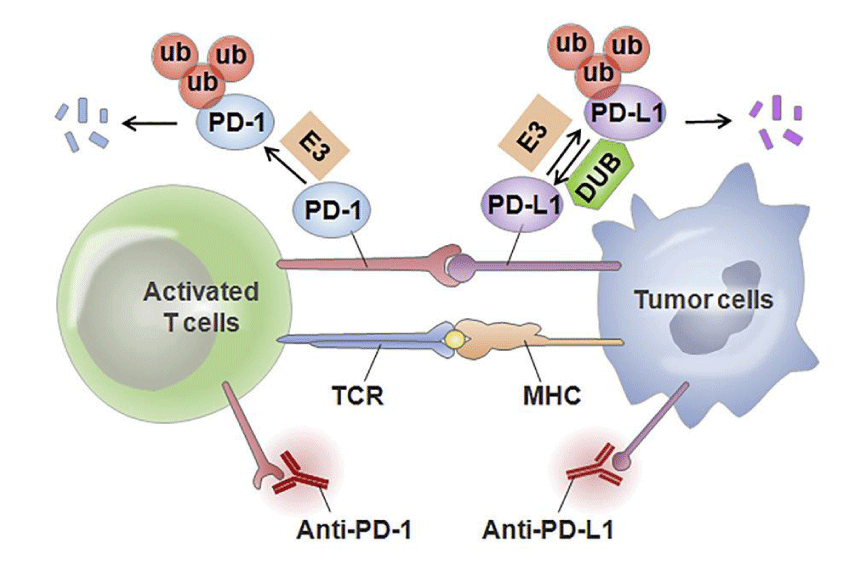
Phosphorylation involves a wide range of regulatory mechanisms of cellular signalling and may affect protein conformation, activity, and interactions [9]. GSK3β, a serine/threonine-protein kinase, was originally thought to be a glycogen degeneration regulator and considered a multifunctional switch in glycosylation. It can mediate direct phosphorylation of various substrates, including eIF2B, Cyclin D1, C-Jun, C-MyC, NFAT, McL-1, etc. [52]. It promotes PD-L1 phosphorylation through the conserved GSK3β phosphorylation motif on PD-L1, and such phosphorylation initiates the interaction between PD-L1 and E3 ligase, which mediates PD-L1 ubiquitination [9]. Ubiquitination is a way to control protein metabolism by controlling proteasome degradation, and proteins are usually tagged with specific polyubiquitin chains before being degraded by proteasome or lysosome pathways [16]. E1 activase, E2 binding enzyme, and E3 ligase mediate ubiquitination with target protein recognition specificity in the proteasome degradation pathway. Among them, there are three E3 ligases targeting PD-L1 protein reported. One is Cullin 3SPOP, which makes PD-L1 ubiquitinated and subsequent degradation. However, Cyclin DCDK4 kinase (a cell cycle kinase) mediates phosphorylation of SPOP, promotes degradation of SPOP by APC/CCdh1, and blocks PD-L1 ubiquitination. It has been proved that Cyclin D-CDK4 kinase can indirectly control the stability of PD-L1 protein [53]. Meanwhile, SPOP deficiency mutations also affect ubiquitin-mediated PD-L1 degradation, resulting in increased PD-L1 levels and reduced tumor-infiltrating lymphocyte numbers in mouse tumors and primary human prostate cancer specimens. The second E3 ligase, β-TrCP, mediates the ubiquitination of the PD-L1 protein that is acidified by GSK3β phosphorous in basal-like breast cancer cells [16].
However, this process is affected by its glycosylation. As mentioned above, glycosylated PD-L1 protein cannot interact with GSK3β to generate phosphorylation. Therefore, glycosylation indirectly prevents ubiquitination and degradation of the PD-L1 protein. The third E3 ligase is STUB1, and the absence of this ligase improves the protein level of PD-L1 in CMTM6 deficient cells. Studies have shown that STUB1 can directly or indirectly reduce the stability of PD-L1 by modifying the lysine of PD-L1 protein in the inner structural domain of cells [54]. The ubiquitination of PD-L1 accelerates the degradation of PD-L1 to achieve the inhibitory effect, while deubiquitination is the reverse process of ubiquitination, usually maintaining the stability of the target protein. Studies have shown that constitutive photomorphogenic signalosome 5(CSN5) COP9 plays a key role in the deubiquitination of PD-L1 and protects PD-L1 from proteasome degradation. Gingerin, an inhibitor of CSN5, can significantly reduce the activity of CSN5 and weaken the stability of PD-L1 protein [21]. The expression of CSN5 is also regulated by TNF-α, one of the inflammatory cytokines secreted by macrophages, which plays an important role in maintaining the immune escape of carcinoma cells. Studies have shown that TNF-α can enhance the stability of PD-L1 protein by activating P65/CSN5. It was shown that CSN5 was indispensable for TNF-α mediated PD-L1 signalling pathway, further emphasizing the role of CSN5 in inhibiting PD-L1 ubiquitination and deubiquitination (Figure 1) [21].
Palmitoylation and acetylation can enhance the stability of PD-L1 protein. Palmitoylation is one of the post-translational modifications of proteins, in which palmitate is covalently linked to most cysteine residues by a thioester bond (also known as S-palmitoylation). Studies have found that palmitoylation of PD-L1 plays an important role in the stability of PD-L1, thus protecting tumor cells from immune surveillance by T cells and destroying palmitoylation of PD-L1 by site-specific mutations or inhibiting the expression of PD-L1 palmitoylation transferase ZDHHC9. Breast cancer cells can be sensitized to T cell killing, inhibiting tumor growth [55]. Acetylation also stabilizes and promotes the expression of PD-L1. Studies have shown a binding site of transcription factor ETS2 in the promoter of PD-L1 and hepatitis B Xinteracting protein in breast cancer cells. HBXIP can upregulate and co-activate transcription factor ETS2 to induce PD-L1 transcription.
Moreover, PD-L1 was induced by binding to acetyltransferase P300 to induce the acetylation of PD-L1 at lys 270 positions (K270), thus stabilizing and accumulating PD-L1 protein in breast cancer cells. It was proved that PD-L1 acetylation is an important way to stabilize PD-L1 protein and a potential direction of targeted therapy research [56]. Pd-l1 binding protein also affects the stability of PD-L1 protein, CKLF like Marvel transmembrane domain-containing protein 6[Chemokinelike factor (CKLF)-like Marvel transmembrane domain-containing family member, CMTM6] is a type III transmembrane protein existing on the cell surface. The absence of STUB1 E3 ubiquitin ligase reversed the level of PD-L1 protein in CMTM6 gene knockout cells, demonstrating that CMTM6 inhibits PD-L1 ubiquitination by binding to PD-L1 protein. By preventing PD-l1 from becoming a lysosomal pathway-mediated degradation target, PD-l1 protein can be protected from degradation, and tumor cells' ability to inhibit T cells can be enhanced by prolonging the half-life of PD-l1 protein (Figure 1) [54,57]. Studies have shown that depletion of CMTM6 reduces the expression of INTERFERon-γ-induced PD-L1 but does not reduce the level of MHC class I on the cell surface and affect antigen presentation, and does not impair the output of PD-L1 from the endoplasmic reticulum and the outward transport of PD-L1 from the Golgi apparatus. It was demonstrated that CMTM6 is not required to transport PD-L1 protein from the er to the cell surface but is required for stable expression of PD-L1 protein on the plasma membrane [57]. In addition, studies have shown that CMTM4, a close family member of CMTM6, also has similar functions to CMTM6 [54]. Another binding protein that affects the stability of PD-L1 is Sigma1, a ligand-regulated whole-membrane chaperone or scaffold protein that contributes to the dynamic balance of cellular proteins and lipids [58].
It has been demonstrated that PD-L1 protein levels are inhibited by RNAi knockdown of Sigma1 and small molecule inhibitors of Sigma1 in TNBC cells expressing Sigma1 and androgen-independent prostate cancer cells. The mechanism is that the Sigma1 regulator can regulate the transport and stability of PD-L1 by regulating the pharmacological reactive protein complex and coordinate with the components of the dynamic balance mechanism of ER protein to regulate the transport of PD-L1 to the plasma membrane through the secretion pathway [58]. Sigma1 inhibitors can isolate PD-L1 protein in the secretion pathway into autophagosomes and induce degradation of PD-L1 through selective autophagy [58]. Pharmacological regulation of Sigma1 can affect cytokines induced by the immune response and regulate the production and activity of PD-L1 through these cytokines mediating extracellular feedback loops or directly binding specific proteins. Its mechanism is the regulation of the tumor immune microenvironment. It was suggested that selective small-molecule Sigma1 ligand could modulate the tumor immune microenvironment [58].
Metabolic reprogramming is considered a marker of tumor and immune development. There is evidence that metabolites can regulate signal transduction through direct interaction or as substrates regulating post-translational modifications of proteins. Therefore, in addition to the supply of bioenergy and biosynthesis, reconnected metabolic pathways also play a role in signal transmission, thereby affecting the expression of PD-L1. - such as interferon-gamma AKTmTORC1 induced by tumor specificity folic acid circulation enzyme 2 folinic acid (methylene dehydrogenase (methylenetetrahydrofolate dehydrogenase 2, MTHFD2) expression, at the same time, MTHFD2 drives the leaf acid cycle to maintain sufficient uridine-related metabolites, including UDPGlcNAc, to promote the O-N-acetylglucosamine glycosylation of global proteins, including C-MYC. Thus, the stability of C-MyC and the transcription level of PD-L1 were enhanced. In pancreatic adenocarcinoma, o-Glcnacylation levels have been positively correlated with MTHFD2 and PD-L1, revealing non-metabolic roles of MTHFD2 in cellular signalling and cancer biology [59]. The subcellular transport process can also indirectly affect the immune escape of tumor cells by affecting the content of PD-L1 protein on the cell surface. Pd-l1 protein is expressed and synthesized in cells and modified and eventually plays a role on the surface of the cell membrane or is secreted out of the cell. However, it may also be transferred from the surface of the cell membrane to the cytoplasm. Studies have shown that PD-L1 protein disappears rapidly after incubation with Quine (an inhibitor of intracellular circulation). Pd-l1 protein and CMTM6 were traced in the intracellular circulating body, indicating that many PD-L1 proteins are constantly metabolized and internalized on the cell membrane. In contrast, PD-L1 in CMTM6 gene knockout cells treated in the same way cannot effectively circulate, as described above. CMTM6, the regulatory factor of PD-L1, can stabilize PD-L1 by inhibiting ubiquitination of PD-L1 protein, and the loss of CMTM6 may lead to degradation of PD-L1 protein in lysosomes after endocytosis [57]. The internalization and release of PD-L1 protein maintain the number of PD-L1 proteins located on the cell membrane as shown in figure 2. It is also an important research idea to inhibit the expression of PD-L1 protein by blocking the internalization cycle pathway. In addition to transport to the cell membrane, PD-L1 can also be transferred from the plasma membrane to the nucleus via endocytosis and cytoplasmic transport. Lys263 at the C-terminal of PD-L1 regulates the expression of pro-inflammatory and immune-resistant genes under the control of p300 and HDAC2-mediated acetylation and deacetylation. These results suggest that transcriptional regulation of PD-L1 may promote immune inflammation in the local tumor microenvironment, making tumors more sensitive to immune checkpoint blocking therapy. Nuclear PD-L1 can also trigger the expression of other immune checkpoint molecules that are not targets of PD-1/PD-L1 blockade and may lead to acquired immunotherapy resistance. Thus, blocking nuclear translocation of PD-L1 with HDAC2 inhibitors may reduce transcription of these immune checkpoint genes, leading to increased CD8+T cell infiltration and decreased TNF-α levels, which in turn enhances the antitumor immune response triggered by therapeutic antibodies to PD-1. It provides a new idea for tumor immunotherapy [60].
Molecular regulatory mechanisms of the microenvironment
Studies have shown that the tumor microenvironment is a dynamic material environment, and cytokines surrounding tumor cells are generated through their secretions and external cell secretions in the process of tumor genesis, which contains nutrients and nutrients a large number of immune cells. In order to meet cellular bioenergy and biosynthesis requirements, tumor cells must adapt to the changing microenvironment. Both tumor cells and immune cells reprogram metabolic pathways to achieve dynamic antagonism. Many specific cytokines and tumor-derived exosomes in TEM can induce PD-L1 expression and initiate immune escape. Pd-l1 mainly acts on the tumor microenvironment rich in lactic acid, while T cell autophagy is induced in the microenvironment deficient in tryptophan, arginine and glucose. In the case of rapid cell growth and lack of surrounding nutrients, anaerobic glucose metabolism increases, resulting in lactic acid accumulation, thus creating the optimal environment for PD-1/PD-L1 interaction and drug resistance of tumor cells [61]. Hypoxia-inducible factor-1α (HIF-1α) is another major cancer driver in the microenvironment. Hypoxia induces hiF-1 α activation and lactic acid accumulation. The binding of HIF-1α to the PD-L1 promoter (a hypoxia response element) stimulates PD-L1 transcription and helps tumour cells escape from the immune system [9]. For example, the enhancer of Zeste homolog 2(EZH2) has been found to regulate the expression of the immunosuppressive molecule PD-L1 in NSCLC cells via HIF-1α. Moreover, the enhancer of EZH2 is an epigenetic regulatory molecule with histone methyltransferase activity, which can promote the formation of an immunosuppressive microenvironment [62]. Figure 3 shows a strong positive correlation between the expression level of TCR and MHC with PD-L1, suggesting that EZH2 can regulate PD-L1 through up-regulation of HIF-1A [62].
Non-codingNon-coding RNAs in tumour cell exosomes can also promote the expression of PD-L1 and induce immune escape. In chronic lymphocytic leukaemia (CLL), monocytes and macrophages have a tumour-promoting phenotype tendency. Including the release of tumour-supporting cytokines and the expression of immunosuppressive molecules such as PD-L1. In addition, CLL-derived exosomes have been shown to induce cancer-related production 2430 · Review · fibroblasts. Studies have shown that non-coding-coding Y RNA hY4 in CLL-derived exosomes can drive the toll-like receptor 7(TLR7) signalling pathway in monocytes and induce PD-L1 expression. Y RNA has also been detected in exosomes from various tumor sources, and tumour-associated slow inflammation and PD-L1 up-regulation have been observed [63]. These findings prove that exosomes in the tumor microenvironment play an important role in promoting tumor progression and are important regulatory factors and inhibitory sites of the tumor microenvironment. Regulation of the microenvironment is a complex and systematic study, and changes in the microenvironment affect T cell infiltration, activation, etc. Research on the immunosuppressive microenvironment is helpful to understand tumor progression and weaken treatment resistance [64].
Small molecule drug therapy and prospects
Under normal physiological conditions, PD-L1 mRNA is subject to strict post-transcriptional regulation. In sharp contrast, a large amount of PD-L1 protein is expressed on the surface of various human tumor cells because cancer cells and other cells in the tumor microenvironment can upregulate THE expression of PD-L1 through interferon-γ upon encountering T cells. Tumor cells and other cells in the tumor microenvironment can also express high levels of PD-L1, and this overexpressed PD-L1 can interact with PD-1 to inhibit immune function. The mechanism of PD-L1's effect on immunosuppression is very complex. Cells expressing PD-L1 can inhibit tumor immunity through a variety of mechanisms. For example, PD-L1+ tumor cells and antigen-presenting cells can induce T cell apoptosis and failure and stimulate human peripheral blood T cells to produce IL-10 for mediated immunosuppression. They can also mediate dendritic cell inhibition and induce T cell differentiation. Pd-l1 can also form a "molecular barrier" on cancer cells, protecting tumor cells from the dissolution of cytotoxic T lymphocytes and effectively preventing immune effector cells from killing cancer cells. Tumor immune escape mediated by the PD-L1 pathway is an "adaptive resistance." Recognizing tumor antigen by Tumor-Infiltrating Lymphocyte (TIL) can initiate this "adaptive resistance." After T cell receptor-specific recognition, TIL releases interferon-γ and may induce these cells to express PD-L1. Pd-l1 does not exist in most normal tissues, but interferon-γ can induce the expression of PD-L1 in almost any nucleated cells [65]. Interferon-γ enhances TIL function by increasing differentiation and stimulating antigen processing and presentation, while PD-L1 binds to its receptor on the cell surface and paralyzes T cells. Therefore, although the physiological function of upregulated PD-L1 expression is to prevent the spread of inflammation and reduce tissue damage, PD-L1 induced in the tumor microenvironment can inhibit the negative feedback regulation of tumor immunity.
Conclusion
In recent years, PD-L1 targeted monoclonal antibodies (MAbs) have been the mainstay of the response to immunosuppression, with pembrolizumab and nivolumab already approved for marketing. It also showed an obvious effect in the treatment of several tumor cells. However, therapeutic antibodies also show some disadvantages, such as limited tissue and tumor penetration; relatively large monoclonal antibodies are difficult to penetrate the complex tumor microenvironment, thus limiting the therapeutic effect of MAb [66,67]. At the same time, a very long half-life, lack of oral bioavailability, immunogenicity, Manufacturing difficulties, and high costs limit the scale and prospects of MAb applications. In addition, monoclonal antibodies against the PD-1/PD-L1 axis are currently only effective in a small number of cases and tumor types [68,69]. Therefore, it is crucial to develop smaller small molecule drugs and improve the specificity of tumor PD-L1 targeting [70-72]. Small molecule drugs may significantly improve the efficacy due to better tumor penetration and oral availability [6]. In the field of molecular structure, the possibility of small molecule drug binding is found through the study of eutectic structure. For example, BMS compounds can bind to the interaction surface of PD-L1 and PD-1, and this interaction between PD-L1 and PD-1 produces an asymmetrical arrangement of opal dimers. There is a small binding site between two protein molecules. Analysis of the interaction between protein molecules and binding inhibitors can characterize the hydrophobic protein-inhibitor contact network, as well as a large number of hydrogen bonds and electrostatic effects of the stable homologous dimer, demonstrating the feasibility of small molecule drug inhibition at the PD-1/PD-L1 immune checkpoint [6]. Recent studies have shown that A compound ARB-272572(compound A) can bind to PD-L1 to form cis-interacting homo-dimer and stabilize PD-L1 homo-dimer to induce rapid internalization PD-L1 on the cell surface by forming A hydrophobic zone between two PD-L1 molecules. Its internalization results in rapid loss of PD-L1 on the cell surface, thus preventing further interaction between PD-L1 and PD-1-expressing cells. However, such internalization is reversible [73-79], meaning that no irreversible damage will be caused to normal cells beyond treatment, which has guiding significance for biological checkpoint and bedside treatment.
Currently, most studies on small molecule inhibitors directly target the binding site of PD-1 and PD-L1 protein, while studies on small molecule inhibitors of the upstream regulatory pathway of PD-L1 are very limited. The regulatory effect of the PD-L1 upstream pathway varies according to different cancer cell types, and differences in the tumor microenvironment will also affect its regulatory effect. Therefore, although small-molecule inhibitors targeting upstream pathways may not be widely applicable, they may also become an adjuvant therapy in cancer treatment.
References
- Escors D, Gato-Cañas M, Zuazo M, Arasanz H, García-Granda MJ, Vera R, Kochan G. The intracellular signalosome of PD-L1 in cancer cells. Signal Transduct Target Ther. 2018 Sep 28;3:26. doi: 10.1038/s41392-018-0022-9. PMID: 30275987; PMCID: PMC6160488.
- Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999 Dec;5(12):1365-9. doi: 10.1038/70932. PMID: 10581077.
- Woods DM, Sodré AL, Villagra A, Sarnaik A, Sotomayor EM, Weber J. HDAC Inhibition Upregulates PD-1 Ligands in Melanoma and Augments Immunotherapy with PD-1 Blockade. Cancer Immunol Res. 2015 Dec;3(12):1375-85. doi: 10.1158/2326-6066.CIR-15-0077-T. Epub 2015 Aug 21. PMID: 26297712; PMCID: PMC4674300.
- M L, P PV, T K, M P, E S, J P, K V W, C L, F C, S D, M SKS, M M, A K, J PI, A S, E S, J W, E M S, A V. Essential role of HDAC6 in the regulation of PD-L1 in melanoma. Mol Oncol. 2016 May;10(5):735-750. doi: 10.1016/j.molonc.2015.12.012. Epub 2016 Jan 6. PMID: 26775640; PMCID: PMC4870131.
- Khan Safir Ullah. Extra Chromosomal Circular DNA: Recent Advances in Research. Journal of Biomedical Research & Environmental Sciences. 2022.
- Zhu H, Bengsch F, Svoronos N, Rutkowski MR, Bitler BG, Allegrezza MJ, Yokoyama Y, Kossenkov AV, Bradner JE, Conejo-Garcia JR, Zhang R. BET Bromodomain Inhibition Promotes Anti-tumor Immunity by Suppressing PD-L1 Expression. Cell Rep. 2016 Sep 13;16(11):2829-2837. doi: 10.1016/j.celrep.2016.08.032. PMID: 27626654; PMCID: PMC5177024.
- Zak KM, Grudnik P, Magiera K, Dömling A, Dubin G, Holak TA. Structural Biology of the Immune Checkpoint Receptor PD-1 and Its Ligands PD-L1/PD-L2. Structure. 2017 Aug 1;25(8):1163-1174. doi: 10.1016/j.str.2017.06.011. PMID: 28768162.
- Pascolutti R, Sun X, Kao J, Maute RL, Ring AM, Bowman GR, Kruse AC. Structure and Dynamics of PD-L1 and an Ultra-High-Affinity PD-1 Receptor Mutant. Structure. 2016 Oct 4;24(10):1719-1728. doi: 10.1016/j.str.2016.06.026. Epub 2016 Sep 8. PMID: 27618663; PMCID: PMC5052120.
- Lázár-Molnár E, Scandiuzzi L, Basu I, Quinn T, Sylvestre E, Palmieri E, Ramagopal UA, Nathenson SG, Guha C, Almo SC. Structure-guided development of a high-affinity human Programmed Cell Death-1: Implications for tumor immunotherapy. EBioMedicine. 2017 Mar;17:30-44. doi: 10.1016/j.ebiom.2017.02.004. Epub 2017 Feb 6. PMID: 28233730; PMCID: PMC5360572.
- Wang Y, Wang H, Yao H, Li C, Fang JY, Xu J. Regulation of PD-L1: Emerging Routes for Targeting Tumor Immune Evasion. Front Pharmacol. 2018 May 22;9:536. doi: 10.3389/fphar.2018.00536. PMID: 29910728; PMCID: PMC5992436.
- Wang W, Li F, Mao Y, Zhou H, Sun J, Li R, Liu C, Chen W, Hua D, Zhang X. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013 Jun;132(6):641-8. doi: 10.1007/s00439-013-1275-6. Epub 2013 Feb 21. PMID: 23430453.
- Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, Grasso CS, Hugo W, Sandoval S, Torrejon DY, Palaskas N, Rodriguez GA, Parisi G, Azhdam A, Chmielowski B, Cherry G, Seja E, Berent-Maoz B, Shintaku IP, Le DT, Pardoll DM, Diaz LA Jr, Tumeh PC, Graeber TG, Lo RS, Comin-Anduix B, Ribas A. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov. 2017 Feb;7(2):188-201. doi: 10.1158/2159-8290.CD-16-1223. Epub 2016 Nov 30. PMID: 27903500; PMCID: PMC5296316.
- George J, Saito M, Tsuta K, Iwakawa R, Shiraishi K, Scheel AH, Uchida S, Watanabe SI, Nishikawa R, Noguchi M, Peifer M, Jang SJ, Petersen I, Büttner R, Harris CC, Yokota J, Thomas RK, Kohno T. Genomic Amplification of CD274 (PD-L1) in Small-Cell Lung Cancer. Clin Cancer Res. 2017 Mar 1;23(5):1220-1226. doi: 10.1158/1078-0432.CCR-16-1069. Epub 2016 Sep 12. PMID: 27620277; PMCID: PMC6329376.
- Clavé S, Pijuan L, Casadevall D, Taus Á, Gimeno J, Hernández-Llodrà S, Rodríguez-Rivera M, Lorenzo M, Menéndez S, Albanell J, Espinet B, Arriola E, Salido M. CD274 (PDL1) and JAK2 genomic amplifications in pulmonary squamous-cell and adenocarcinoma patients. Histopathology. 2018 Jan;72(2):259-269. doi: 10.1111/his.13339. Epub 2017 Oct 27. PMID: 28795418.
- Ikeda S, Okamoto T, Okano S, Umemoto Y, Tagawa T, Morodomi Y, Kohno M, Shimamatsu S, Kitahara H, Suzuki Y, Fujishita T, Maehara Y. PD-L1 Is Upregulated by Simultaneous Amplification of the PD-L1 and JAK2 Genes in Non-Small Cell Lung Cancer. J Thorac Oncol. 2016 Jan;11(1):62-71. doi: 10.1016/j.jtho.2015.09.010. PMID: 26762740.
- Kress TR, Sabò A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer. 2015 Oct;15(10):593-607. doi: 10.1038/nrc3984. Epub 2015 Sep 18. PMID: 26383138.
- Shi Y. Regulatory mechanisms of PD-L1 expression in cancer cells. Cancer Immunol Immunother. 2018 Oct;67(10):1481-1489. doi: 10.1007/s00262-018-2226-9. Epub 2018 Aug 17. PMID: 30120503.
- Khan SU, Khan MU. The mechanism of mammalian mitochondrial quality control system. Journal of Chemistry and Nutritional Biochemistry. 2021;59-69. doi: 10.48185/jcnb.v2i2.387.
- KATAOKA K, SHIRAISHI Y, TAKEDA Y, Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016:534(7607):402-406.
- Green MR, Rodig S, Juszczynski P, Ouyang J, Sinha P, O'Donnell E, Neuberg D, Shipp MA. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res. 2012 Mar 15;18(6):1611-8. doi: 10.1158/1078-0432.CCR-11-1942. Epub 2012 Jan 23. Erratum in: Clin Cancer Res. 2012 Apr 1;18(7):2117. PMID: 22271878; PMCID: PMC3321508.
- Fang W, Zhang J, Hong S, Zhan J, Chen N, Qin T, Tang Y, Zhang Y, Kang S, Zhou T, Wu X, Liang W, Hu Z, Ma Y, Zhao Y, Tian Y, Yang Y, Xue C, Yan Y, Hou X, Huang P, Huang Y, Zhao H, Zhang L. EBV-driven LMP1 and IFN-γ up-regulate PD-L1 in nasopharyngeal carcinoma: Implications for oncotargeted therapy. Oncotarget. 2014 Dec 15;5(23):12189-202. doi: 10.18632/oncotarget.2608. PMID: 25361008; PMCID: PMC4322961.
- Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, Parisi G, Saus CP, Torrejon DY, Graeber TG, Comin-Anduix B, Hu-Lieskovan S, Damoiseaux R, Lo RS, Ribas A. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017 May 9;19(6):1189-1201. doi: 10.1016/j.celrep.2017.04.031. Erratum in: Cell Rep. 2019 Dec 10;29(11):3766. PMID: 28494868; PMCID: PMC6420824.
- Khan SU, Khan MU, Kalsoom F, Khan MI, Gao S, Unar A, Zubair M, Bilal M. Mechanisms of gene regulation by histone degradation in adaptation of yeast: an overview of recent advances. Arch Microbiol. 2022 Apr 28;204(5):287. doi: 10.1007/s00203-022-02897-8. PMID: 35482104.
- Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, Chang SS, Lin WC, Hsu JM, Hsu YH, Kim T, Chang WC, Hsu JL, Yamaguchi H, Ding Q, Wang Y, Yang Y, Chen CH, Sahin AA, Yu D, Hortobagyi GN, Hung MC. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell. 2016 Dec 12;30(6):925-939. doi: 10.1016/j.ccell.2016.10.010. Epub 2016 Nov 17. PMID: 27866850; PMCID: PMC5171205.
- Wang X, Yang L, Huang F, Zhang Q, Liu S, Ma L, You Z. Inflammatory cytokines IL-17 and TNF-α up-regulate PD-L1 expression in human prostate and colon cancer cells. Immunol Lett. 2017 Apr;184:7-14. doi: 10.1016/j.imlet.2017.02.006. Epub 2017 Feb 14. PMID: 28223102; PMCID: PMC5362328.
- Marzec M, Zhang Q, Goradia A, Raghunath PN, Liu X, Paessler M, Wang HY, Wysocka M, Cheng M, Ruggeri BA, Wasik MA. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc Natl Acad Sci U S A. 2008 Dec 30;105(52):20852-7. doi: 10.1073/pnas.0810958105. Epub 2008 Dec 16. PMID: 19088198; PMCID: PMC2634900.
- Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI, Park YM, Oh S, Shin JG, Yao S, Chen L, Choi IH. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274). FEBS Lett. 2006 Feb 6;580(3):755-62. doi: 10.1016/j.febslet.2005.12.093. Epub 2006 Jan 9. PMID: 16413538.
- Li N, Wang J, Zhang N, Zhuang M, Zong Z, Zou J, Li G, Wang X, Zhou H, Zhang L, Shi Y. Cross-talk between TNF-α and IFN-γ signaling in induction of B7-H1 expression in hepatocellular carcinoma cells. Cancer Immunol Immunother. 2018 Feb;67(2):271-283. doi: 10.1007/s00262-017-2086-8. Epub 2017 Oct 31. PMID: 29090321.
- Concha-Benavente F, Srivastava R M, Trivedi S. Identification of the cell-intrinsic and -extrinsic pathways downstream of EGFR and IFNgamma that induce PD-L1 expres-sion in head and neck cancer. Cancer Res. 2016:76(5):1031-43.
- Khan S, Khan M. Molecular developments in cell models of fatty liver disease. DYSONALife Science. 2022;1:16-29. doi: 10.30493/DLS.2022.325915.
- Moon JW, Kong SK, Kim BS, Kim HJ, Lim H, Noh K, Kim Y, Choi JW, Lee JH, Kim YS. IFNγ induces PD-L1 overexpression by JAK2/STAT1/IRF-1 signaling in EBV-positive gastric carcinoma. Sci Rep. 2017 Dec 19;7(1):17810. doi: 10.1038/s41598-017-18132-0. PMID: 29259270; PMCID: PMC5736657.
- Liu J, Hamrouni A, Wolowiec D, Coiteux V, Kuliczkowski K, Hetuin D, Saudemont A, Quesnel B. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-{gamma} and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood. 2007 Jul 1;110(1):296-304. doi: 10.1182/blood-2006-10-051482. Epub 2007 Mar 15. PMID: 17363736.
- Okita R, Maeda A, Shimizu K, Nojima Y, Saisho S, Nakata M. PD-L1 overexpression is partially regulated by EGFR/HER2 signaling and associated with poor prognosis in patients with non-small-cell lung cancer. Cancer Immunol Immunother. 2017 Jul;66(7):865-876. doi: 10.1007/s00262-017-1986-y. Epub 2017 Mar 25. PMID: 28341875.
- Khan SU. Therapeutic application of genetically engineered ribosome-inactivating toxin proteins for cancer. J Biomed Res Environ Sci. 2021;2(12):1216-1228. doi:10.37871/jbres1375.
- Fiedler M, Schulz D, Piendl G, Brockhoff G, Eichberger J, Menevse AN, Beckhove P, Hautmann M, Reichert TE, Ettl T, Bauer RJ. Buparlisib modulates PD-L1 expression in head and neck squamous cell carcinoma cell lines. Exp Cell Res. 2020 Nov 1;396(1):112259. doi: 10.1016/j.yexcr.2020.112259. Epub 2020 Sep 6. PMID: 32898555.
- Sumimoto H, Takano A, Teramoto K, Daigo Y. RAS-Mitogen-Activated Protein Kinase Signal Is Required for Enhanced PD-L1 Expression in Human Lung Cancers. PLoS One. 2016 Nov 15;11(11):e0166626. doi: 10.1371/journal.pone.0166626. PMID: 27846317; PMCID: PMC5112979.
- Khan SU, Khan MU. Review on gene regulation: DNA-protein and protein-protein interactions and their regulatory elements. Journal of Chemistry and Nutritional Biochemistry. 2021;2(2):35-45. doi: 10.48185/jcnb.v2i2.378.
- Wu Y, Zhang C, Liu X, He Z, Shan B, Zeng Q, Zhao Q, Zhu H, Liao H, Cen X, Xu X, Zhang M, Hou T, Wang Z, Yan H, Yang S, Sun Y, Chen Y, Wu R, Xie T, Chen W, Najafov A, Ying S, Xia H. ARIH1 signaling promotes anti-tumor immunity by targeting PD-L1 for proteasomal degradation. Nat Commun. 2021 Apr 20;12(1):2346. doi: 10.1038/s41467-021-22467-8. PMID: 33879767; PMCID: PMC8058344.
- Yang L, Huang F, Mei J, Wang X, Zhang Q, Wang H, Xi M, You Z. Posttranscriptional Control of PD-L1 Expression by 17β-Estradiol via PI3K/Akt Signaling Pathway in ERα-Positive Cancer Cell Lines. Int J Gynecol Cancer. 2017 Feb;27(2):196-205. doi: 10.1097/IGC.0000000000000875. PMID: 27870715; PMCID: PMC5258765.
- Quandt D, Jasinski-Bergner S, Müller U, Schulze B, Seliger B. Synergistic effects of IL-4 and TNFα on the induction of B7-H1 in renal cell carcinoma cells inhibiting allogeneic T cell proliferation. J Transl Med. 2014 May 30;12:151. doi: 10.1186/1479-5876-12-151. PMID: 24885059; PMCID: PMC4079621.
- Khan SU, Khan MU. Treatment of diabetic muscular hyperplasia with natural and nutritional supplements. Global Journal of Biotechnology and Biomaterial Science. 2022;8(1):001-008.
- Boes M, Meyer-Wentrup F. TLR3 triggering regulates PD-L1 (CD274) expression in human neuroblastoma cells. Cancer Lett. 2015 May 28;361(1):49-56. doi: 10.1016/j.canlet.2015.02.027. Epub 2015 Feb 16. PMID: 25697485.
- Coelho MA, de Carné Trécesson S, Rana S, Zecchin D, Moore C, Molina-Arcas M, East P, Spencer-Dene B, Nye E, Barnouin K, Snijders AP, Lai WS, Blackshear PJ, Downward J. Oncogenic RAS Signaling Promotes Tumor Immunoresistance by Stabilizing PD-L1 mRNA. Immunity. 2017 Dec 19;47(6):1083-1099.e6. doi: 10.1016/j.immuni.2017.11.016. Epub 2017 Dec 12. PMID: 29246442; PMCID: PMC5746170.
- Mao R, Tan X, Xiao Y, Wang X, Wei Z, Wang J, Wang X, Zhou H, Zhang L, Shi Y. Ubiquitin C-terminal hydrolase L1 promotes expression of programmed cell death-ligand 1 in non-small-cell lung cancer cells. Cancer Sci. 2020 Sep;111(9):3174-3183. doi: 10.1111/cas.14529. Epub 2020 Jul 6. PMID: 32539182; PMCID: PMC7469845.
- Qin G, Wang X, Ye S. NPM1 upregulates the transcrip-tion of PD-L1 and suppresses T cell activity in triple-negative breast cancer. Nat Commun. 2020;11(1):1669.
- Chen XM. MicroRNA signatures in liver diseases. World J Gastroenterol. 2009 Apr 14;15(14):1665-72. doi: 10.3748/wjg.15.1665. PMID: 19360909; PMCID: PMC2668771.
- Gong AY, Zhou R, Hu G, Li X, Splinter PL, O'Hara SP, LaRusso NF, Soukup GA, Dong H, Chen XM. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J Immunol. 2009 Feb 1;182(3):1325-33. doi: 10.4049/jimmunol.182.3.1325. PMID: 19155478; PMCID: PMC2652126.
- Wang X, Li J, Dong K, Lin F, Long M, Ouyang Y, Wei J, Chen X, Weng Y, He T, Zhang H. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015 Mar;27(3):443-52. doi: 10.1016/j.cellsig.2014.12.003. Epub 2014 Dec 10. PMID: 25499621.
- Khan SU, Khan MU. Recent Developments and Applications of Single-Cell RNA Sequencing Technology in Cell Classification. J Biomed Res Environ Sci. 2021 Dec 29;2(12):1283-1290. doi: 10.37871/jbres1383.
- Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Araujo L, Carbone DP, Shilo K, Giri DK, Kelnar K, Martin D, Komaki R, Gomez DR, Krishnan S, Calin GA, Bader AG, Welsh JW. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst. 2015 Nov 17;108(1):djv303. doi: 10.1093/jnci/djv303. PMID: 26577528; PMCID: PMC4862407.
- Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, Diao L, Wang J, Roybal J, Patel M, Ungewiss C, Peng D, Antonia S, Mediavilla-Varela M, Robertson G, Suraokar M, Welsh JW, Erez B, Wistuba II, Chen L, Peng D, Wang S, Ullrich SE, Heymach JV, Kurie JM, Qin FX. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014 Oct 28;5:5241. doi: 10.1038/ncomms6241. PMID: 25348003; PMCID: PMC4212319.
- Khan SU, Khan MU. The role of amino acid metabolic reprogramming in tumor development and immunotherapy. Biochemistry and Molecular Biology. 2022;7(1):6-12. doi: 10.11648/j.bmb.20220701.12.
- Fujita Y, Yagishita S, Hagiwara K, Yoshioka Y, Kosaka N, Takeshita F, Fujiwara T, Tsuta K, Nokihara H, Tamura T, Asamura H, Kawaishi M, Kuwano K, Ochiya T. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther. 2015 Apr;23(4):717-27. doi: 10.1038/mt.2015.10. Epub 2015 Jan 19. PMID: 25597412; PMCID: PMC4395779.
- Shao L, He Q, Wang J, He F, Lin S, Wu L, Gao Y, Ma W, Dong J, Yang X, Li F. MicroRNA-326 attenuates immune escape and prevents metastasis in lung adenocarcinoma by targeting PD-L1 and B7-H3. Cell Death Discov. 2021 Jun 15;7(1):145. doi: 10.1038/s41420-021-00527-8. PMID: 34131111; PMCID: PMC8206349.
- Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, Song W, Chen Y, OuYang J, Chen J, Kong F, Dong Y, Jiang SW, Li W, Wang P, Yuan Z, Wan X, Wang C, Li W, Zhang X, Chen K. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. 2016 May 5;7:11406. doi: 10.1038/ncomms11406. PMID: 27147225; PMCID: PMC4858750.
- Zhao L, Yu H, Yi S, Peng X, Su P, Xiao Z, Liu R, Tang A, Li X, Liu F, Shen S. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget. 2016 Jul 19;7(29):45370-45384. doi: 10.18632/oncotarget.9659. PMID: 27248318; PMCID: PMC5216728.
- AUDRITO V, SERRA S, STINGI A, PD-L1 up-regulation in melanoma increases disease aggressiveness and is mediated through miR-17-5p .Oncotarget. 2017;8(9):15894-911.
- Khan SU, Khan MU. Advances in innate immune memory of macrophages. Explor Immunol. 2022;2:428–41. doi: 10.37349/ei.2022.00060.
- Cioffi M, Trabulo SM, Vallespinos M, Raj D, Kheir TB, Lin ML, Begum J, Baker AM, Amgheib A, Saif J, Perez M, Soriano J, Desco M, Gomez-Gaviro MV, Cusso L, Megias D, Aicher A, Heeschen C. The miR-25-93-106b cluster regulates tumor metastasis and immune evasion via modulation of CXCL12 and PD-L1. Oncotarget. 2017 Mar 28;8(13):21609-21625. doi: 10.18632/oncotarget.15450. PMID: 28423491; PMCID: PMC5400610.
- Lastwika KJ, Wilson W 3rd, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, Liu LN, Gills JJ, Dennis PA. Control of PD-L1 Expression by Oncogenic Activation of the AKT-mTOR Pathway in Non-Small Cell Lung Cancer. Cancer Res. 2016 Jan 15;76(2):227-38. doi: 10.1158/0008-5472.CAN-14-3362. Epub 2015 Dec 4. PMID: 26637667.
- Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, Khoo KH, Chang SS, Cha JH, Kim T, Hsu JL, Wu Y, Hsu JM, Yamaguchi H, Ding Q, Wang Y, Yao J, Lee CC, Wu HJ, Sahin AA, Allison JP, Yu D, Hortobagyi GN, Hung MC. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016 Aug 30;7:12632. doi: 10.1038/ncomms12632. PMID: 27572267; PMCID: PMC5013604.
- McCubrey JA, Steelman LS, Bertrand FE, Davis NM, Sokolosky M, Abrams SL, Montalto G, D'Assoro AB, Libra M, Nicoletti F, Maestro R, Basecke J, Rakus D, Gizak A, Demidenko ZN, Cocco L, Martelli AM, Cervello M. GSK-3 as potential target for therapeutic intervention in cancer. Oncotarget. 2014 May 30;5(10):2881-911. doi: 10.18632/oncotarget.2037. PMID: 24931005; PMCID: PMC4102778.
- Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, Tan Y, Ci Y, Wu F, Dai X, Guo J, Huang YH, Fan C, Ren S, Sun Y, Freeman GJ, Sicinski P, Wei W. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018 Jan 4;553(7686):91-95. doi: 10.1038/nature25015. Epub 2017 Nov 16. Erratum in: Nature. 2019 Jul;571(7766):E10. PMID: 29160310; PMCID: PMC5754234.
- Mezzadra R, Sun C, Jae LT. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549(7670):106-110.
- Yang Y, Hsu JM, Sun L, Chan LC, Li CW, Hsu JL, Wei Y, Xia W, Hou J, Qiu Y, Hung MC. Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res. 2019 Jan;29(1):83-86. doi: 10.1038/s41422-018-0124-5. Epub 2018 Dec 4. PMID: 30514902; PMCID: PMC6318320.
- Xu FF, Sun HM, Fang RP, Zhang L, Shi H, Wang X, Fu XL, Li XM, Shi XH, Wu Y, Ye K, Zhang WY, Ye LH. The modulation of PD-L1 induced by the oncogenic HBXIP for breast cancer growth. Acta Pharmacol Sin. 2022 Feb;43(2):429-445. doi: 10.1038/s41401-021-00631-6. Epub 2021 Apr 6. PMID: 33824459; PMCID: PMC8791967.
- Burr ML, Sparbier CE, Chan YC. CMTM6 main-tains the expression of PD-L1 and regulates anti-tumour immu-nity. Nature. 2017:549(7670):101-5.
- Maher CM, Thomas JD, Haas DA, Longen CG, Oyer HM, Tong JY, Kim FJ. Small-Molecule Sigma1 Modulator Induces Autophagic Degradation of PD-L1. Mol Cancer Res. 2018 Feb;16(2):243-255. doi: 10.1158/1541-7786.MCR-17-0166. Epub 2017 Nov 8. PMID: 29117944.
- Shang M, Yang H, Yang R, Chen T, Fu Y, Li Y, Fang X, Zhang K, Zhang J, Li H, Cao X, Gu J, Xiao J, Zhang Q, Liu X, Yu Q, Wang T. The folate cycle enzyme MTHFD2 induces cancer immune evasion through PD-L1 up-regulation. Nat Commun. 2021 Mar 29;12(1):1940. doi: 10.1038/s41467-021-22173-5. PMID: 33782411; PMCID: PMC8007798.
- Gao Y, Nihira NT, Bu X, Chu C, Zhang J, Kolodziejczyk A, Fan Y, Chan NT, Ma L, Liu J, Wang D, Dai X, Liu H, Ono M, Nakanishi A, Inuzuka H, North BJ, Huang YH, Sharma S, Geng Y, Xu W, Liu XS, Li L, Miki Y, Sicinski P, Freeman GJ, Wei W. Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat Cell Biol. 2020 Sep;22(9):1064-1075. doi: 10.1038/s41556-020-0562-4. Epub 2020 Aug 24. PMID: 32839551; PMCID: PMC7484128.
- Robainas M, Otano R, Bueno S, Ait-Oudhia S. Understanding the role of PD-L1/PD1 pathway blockade and autophagy in cancer therapy. Onco Targets Ther. 2017 Mar 23;10:1803-1807. doi: 10.2147/OTT.S132508. PMID: 28367063; PMCID: PMC5370068.
- Zhao Y, Wang XX, Wu W, Long H, Huang J, Wang Z, Li T, Tang S, Zhu B, Chen D. EZH2 regulates PD-L1 expression via HIF-1α in non-small cell lung cancer cells. Biochem Biophys Res Commun. 2019 Sep 17;517(2):201-209. doi: 10.1016/j.bbrc.2019.07.039. Epub 2019 Jul 19. PMID: 31331645.
- Haderk F, Schulz R, Iskar M, Cid LL, Worst T, Willmund KV, Schulz A, Warnken U, Seiler J, Benner A, Nessling M, Zenz T, Göbel M, Dürig J, Diederichs S, Paggetti J, Moussay E, Stilgenbauer S, Zapatka M, Lichter P, Seiffert M. Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci Immunol. 2017 Jul 28;2(13):eaah5509. doi: 10.1126/sciimmunol.aah5509. PMID: 28754746.
- Wei Y, Xiao X, Lao XM, Zheng L, Kuang DM. Immune landscape and therapeutic strategies: new insights into PD-L1 in tumors. Cell Mol Life Sci. 2021 Feb;78(3):867-887. doi: 10.1007/s00018-020-03637-1. Epub 2020 Sep 17. PMID: 32940722.
- Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015 Sep;125(9):3384-91. doi: 10.1172/JCI80011. Epub 2015 Sep 1. PMID: 26325035; PMCID: PMC4588282.
- Lee CM, Tannock IF. The distribution of the therapeutic monoclonal antibodies cetuximab and trastuzumab within solid tumors. BMC Cancer. 2010 Jun 3;10:255. doi: 10.1186/1471-2407-10-255. PMID: 20525277; PMCID: PMC2889896.
- Zarganes-Tzitzikas T, Konstantinidou M, Gao Y, Krzemien D, Zak K, Dubin G, Holak TA, Dömling A. Inhibitors of programmed cell death 1 (PD-1): a patent review (2010-2015). Expert Opin Ther Pat. 2016 Sep;26(9):973-7. doi: 10.1080/13543776.2016.1206527. Epub 2016 Jul 6. PMID: 27367741; PMCID: PMC5379636.
- Tan S, Zhang CW, Gao GF. Seeing is believing: anti-PD-1/PD-L1 monoclonal antibodies in action for checkpoint blockade tumor immunotherapy. Signal Transduct Target Ther. 2016 Nov 25;1:16029. doi: 10.1038/sigtrans.2016.29. PMID: 29263905; PMCID: PMC5661648.
- Park JJ, Thi EP, Carpio VH. Checkpoint inhibition through small molecule-induced internalization of programmed death-ligand 1. Nat Commun. 2021;12(1):1222.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.