> Medicine Group. 2020 Oct 31;1(6):256-262. doi: 10.37871/jbres1152.
Association between Signal Transducer and Activator of Transcription 4 Genetic Polymorphisms and the Spontaneous Clearance of Hepatitis B Surface Antigen: A Large Population Case Control Study in China
Xun Qi1,3#, Qirong Jiang1#, Ying Lv3, Sisi Yang1, Jing Li1, Yuxian Huang1,3, Liang Chen3 and Jiming Zhang1,2*
2Key laboratory of Medical Molecular Virology of the Ministries of Education and Health (MOH & MOE), Fudan University, Shanghai, China
3Department of Hepatology, Shanghai Public Health Clinical Center, Fudan University, Shanghai, China
#Xun Qi and Qirong Jiang contributed equally to the work
- Chronic hepatitis B
- HBsAg spontaneous clearance
- Single nucleotide polymorphism
- Signal transducer and activator of transcription 4
Aim: Several host factors mediating immune response influence susceptibility to Hepatitis B Virus (HBV) infection, ability to clear the virus, and maintenance of a chronic state. Signal Transducer and Activator of Transcription 4 (STAT4) variations are correlated with the risk of developing autoimmune diseases. However, there have been few studies to assess the relationship between STAT4 variations and Hepatitis B surface Antigen (HBsAg) clearance in adults infected with HBV. Our aim was to evaluate the association between genetic variants in STAT4 and HBsAg clearance in a large sample size population.
Methods: This case control study included Chronic Hepatitis B (CHB) (n = 1.688), HBsAg Clearance after Treatment (TC) (n = 170), HBV Uninfected (HC) (n = 1.012), and HBsAg Spontaneous Clearance (SC) (n = 1,052) patients. In the CHB group, patients were categorized into four subgroups: the Immune Tolerant (IT), Immune Active (IA), Inactive (IC), and Immune Reactivation (IR) phases, with 97, 855, 198, and 538 patients in each subgroup, respectively.
Results: We found that the G allele in STAT4 rs7574865 was more frequent in the CHB and TC groups, compared with the SC group, whereas the STAT4 rs7574865 GG genotype was more frequent in the CHB and TC group, compared with the SC group in the dominant model. However, there was no statistical significance in genotype between TC and CHB, nor between the IT, IA, IC, and IR groups.
Conclusions: The prevalence of the minor allele rs7574865 T was higher in subjects with spontaneously cleared HBV infections than in CHB patients.
Hepatitis B Virus (HBV) infection is a major global health problem, though the effective HBV vaccine is widely used. HBV infects about 2.5 billion people worldwide and 300 million of those are chronically infected [1,2]. China has one of the highest HBV carrier prevalences in the world. It is estimated that there are approximately 30 million Chronic HBV (CHB) patients in China, almost 5% - 6% of the entire population [3]. About 95% of adults can successfully clear HBV after being infected and 5% - 10% will develop CHB; even fewer progress to liver failure [4]. Several host and viral genetic factors are known to influence susceptibility to HBV infection, ability to clear the virus, and maintenance of a chronic state. Among these factors, host immunogenetic background has a significant role in mediating the clinical progression of viral hepatitis infections [5-8].
Signal Transducer and Activator of Transcription 4 (STAT4), a member of the STAT family, can be activated by several cytokines including IL 12, type I Interferon (IFN), and IL 23, as well as lead to T helper (Th) type 1 and 17 differentiation, monocyte activation, and production of IFN-γ [9]. In fact, more and more studies indicated that STAT4 variations are correlated with the risk of developing autoimmune diseases. The Single Nucleotide Polymorphism (SNP) rs7574865, which is in the third intron of the STAT4 gene, is associated with a higher risk of rheumatoid arthritis [10-13], Systemic Lupus Erythematosus (SLE) [14], type I diabetes [15], and systemic scleroderma [16]. Furthermore, it is reported that the G allele of rs7574865 can increase the risk of Hepatocellular Carcinoma (HCC) and decrease the efficacy of IFN treatment in CHB patients [17-19].
However, few studies have assessed the relationship between STAT4 variations and Hepatitis B surface Antigen (HBsAg) clearance in adults infected with HBV, and the results of these studies have been inconsistent. Our aim was to evaluate the association between the genetic variations in STAT4 and HBsAg clearance in HBV infected adults in a large sample size population.
Patients
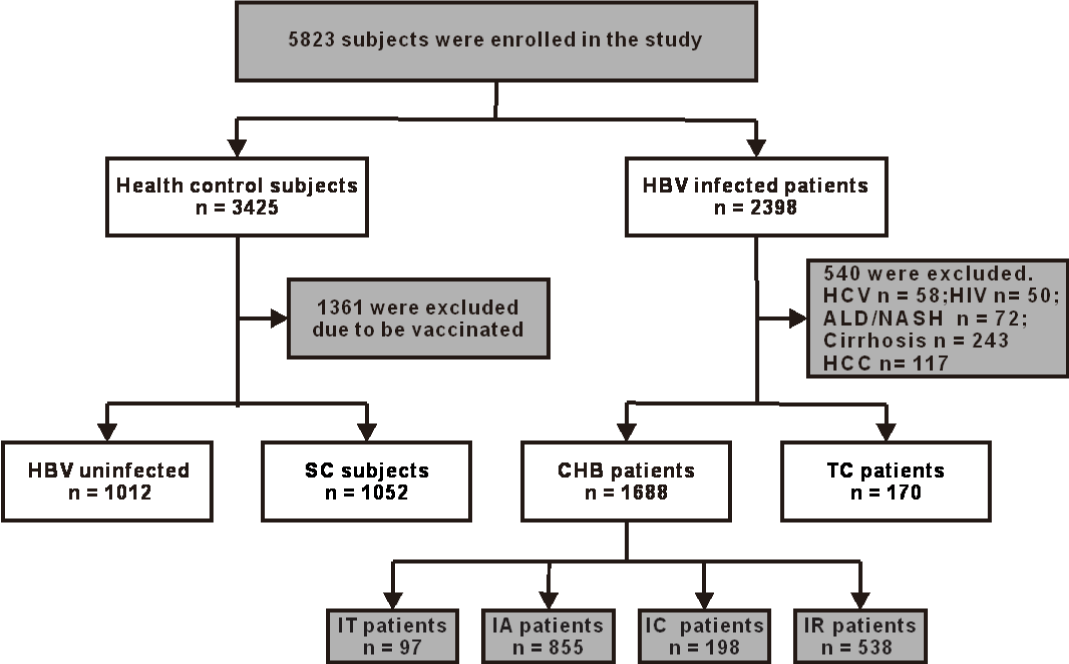
This case control study enrolled 1,858 of 2,398 patients with CHB infection, aged 18-70 years, who were followed up at our hospitals between March 2016 and December 2018. These patients were divided into two groups: CHB patients, and HBsAg Clearance after Treatment (TC). There were 1,688 and 170 patients in each group, respectively. In the CHB group, the patients were categorized into four subgroups according to the natural history [20]: immune Tolerant Phase (IT), Immune Active Phase (IA), Inactive Phase (IC), and Immune Reactivation Phase (IR) with 97, 855, 198, and 538 patients in each subgroup, respectively. TC was defined as CHB patients who received antiviral treatment for at least 1 year and had HBsAg clearance (Figure 1).
Major inclusion criteria for the CHB group included HBV monoinfection with detectable HBsAg at screening and for at least 6 months prior to the onset of the study. The evidence of cirrhosis or HCC by imaging, or a history of alcohol abuse were excluded.
Of the 3,425 healthy subjects, 2,064 were enrolled into this study as controls. These subjects were divided into the HBV uninfected (HC) group and HBsAg Spontaneous Clearance (SC) group with 1,012 and 1,052 individuals in each group, respectively. The HC group was defined as those in which all HBV markers were negative. SC subjects were defined as those who were negative for HBsAg, but positive for both the Hepatitis B surface Antibody (HBsAb) and Hepatitis B core Antibody (HBcAb). All control subjects had no other ailments and had normal liver function (Figure 1).
The study was in complied with the Helsinki Declaration. Our Center’s Ethics Committee reviewed and approved the study design (KY2016-196) and all patients provided written informed consent.
Study design
All data of the subjects were collected at the baseline, including history, liver function, and White Blood Cell Count (WBC), Platelet Count (PLT), and HBV markers. In CHB infected patients, HBV DNA load and HBV genotype were tested.
DNA isolation and rs7574865 genotyping
Genomic DNA was extracted from 1 ml of peripheral whole blood from each subject using Qiagen DNA kits (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The STAT4 variants were genotyped using a real-time Polymerase Chain Reaction (PCR) in a 50 μl reaction system including 20ng of genomic DNA. The thermal cycling conditions applied were initial activation for 5 min at 95°C, 40 cycles of denaturation at 94°C for 30s, annealing at 57°C for 30s, and extension at 72°C for 30s. All the samples were analyzed by direct sequencing.
Statistical methods
All data were processed using Stata 16.0 software (Stata Corporation, College Station, TX, USA). A two-tailed p < 0.05 was considered statistically significant. Continuous variables were expressed as median (range) and categorical variables were expressed as number (%). Continuous variables were analyzed using the Student’s t-test or Wilcoxon test. Categorical variables were analyzed using the chi-square test or Fisher’s exact test. The logistic regression model was used to estimate the univariate and multivariate Odds Ratios (OR) of various factors related to HBsAg clearance after HBV infection.
Baseline characteristics of the subjects
In this study, 5,823 subjects were screened, and 3,922 met the inclusion criteria and were included (Figure 1). The proportion of male, age, and the values of Alanine Transaminase (ALT) and Aspartate Aminotransferase (AST) were higher in the CHB and TC groups than in the HC and SC groups. However, the values of WBC and PLT in the CHB and TC groups were lower than in the HC and SC groups. In CHB patients, the major HBV genotype was the C genotype, and the percentage of Hepatitis B e Antigen (HBeAg) positive patients was 56% (Table 1).
| Table 1: The characteristics of the subjects in the study. | ||||
| HC | SC | CHB | TC | |
| Total (n) | 1.012 | 1.052 | 1.688 | 170 |
| Male (%) | 548 (54%) | 548 (52%) | 1,097 (65%) | 132 (78%) |
| Age (years) | 31 (18 - 69) | 36 (20 - 65) | 45 (18 - 68) | 43 (24 - 65) |
| ALT (U/L) | 20 (4 - 43) | 20 (5 - 38) | 47 (6 - 1586) | 43 (9 - 112) |
| AST (U/L) | 20 (9 - 37) | 20 (6 - 31) | 37 (11 - 899) | 33 (12 - 117) |
| TBil (μmol/L) | 16 (2-21) | 16 (2-29) | 22 (3-40) | 20 (6 - 38) |
| ALB (g/L) | 47 (36 - 54) | 47 (39 - 57) | 45 (36 - 54) | 46 (40 - 55) |
| WBC (×109/L) | 6.65 (3.83 - 9.92) | 6.63 (3.97 - 9.53) | 5.66 (3.61 - 9.90) | 5.01 (2.41 - 9.40) |
| PLT (×1012/L) | 244 (116 - 451) | 241 (111 - 447) | 177 (91 - 375) | 172 (77 - 378) |
| HBV genotype (B/C/unknown) | - | - | 371/632/685 (22%/37%/41%) |
- |
| HBsAg (log10IU/ml) | - | - | 4.02 (1.23 - 4.72) | - |
| HBeAg positive (%) | - | - | 952 (56%) | - |
| HBV DNA (log10IU/ml) | - | - | 7.05 (0.00 - 9.46) | - |
| Values are expressed as median (range) or number of subjects (%). HC: HBV Uninfected; HBsAg: Hepatitis B Surface Antigen; HBeAg: Hepatitis B e-Antigen; SC: Hepatitis B Surface Antigen (Hbsag) Spontaneous Clearance; CHB: Chronic Hepatitis B; TC: HBsAg Clearance after Treatment; ALT: Alanine Aminotransferase; AST: Aspartate Aminotransferase; TBil: Total Bilirubin; ALB: Albumin; WB: White Blood Cell Count; PLT: Platelet Count; HBV: Hepatitis B Virus; HBsAg: Hepatitis B Surface Antigen; HBeAg: Hepatitis B e-Antigen. |
||||
Association between STAT4 polymorphisms and HBsAg clearance
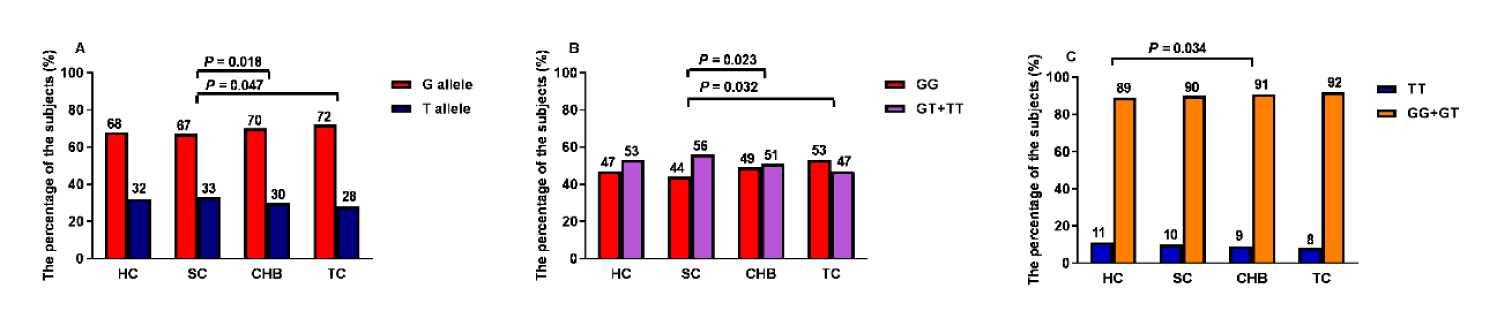
To evaluate the relationship between rs7574865 variation and HBsAg spontaneous clearance, the SC group was compared with the CHB, TC, and HC groups. The frequency of the G allele in STAT4 rs7574865 was higher in the CHB and TC groups, compared with the SC group (CHB vs. SC: 70% vs. 67%, p = 0.018; TC vs. SC: 72% vs. 67%, p = 0.047) (Figure 2A). However, there was no significant difference between the HC and CHB group (Supplementary table 1).
The similar trend was observed in the dominant model. The STAT4 rs7574865 GG genotype was more frequent in the CHB and TC groups, compared with the SC group (CHB vs. SC: 49% vs. 44%, p = 0.023; TC vs. SC: 53% vs. 44%, p = 0.032; Figure 2B). However, there was no significant difference between the HC and CHB groups (Supplementary table 1), and no statistical significance in the recession model between the CHB, TC, and SC groups (Figure 2C).
Furthermore, the association between rs7574865 variation and HBsAg clearance after the treatment was analyzed, which indicated no statistical significance regarding the genotype between the TC and CHB groups (G allele: 72% vs. 70%, p = 0.359; the dominant model: 53% vs. 49%, p = 0.272; the recession model: 8% vs. 9%, p = 0.857; figure 2).
Association between STAT4 polymorphisms and HBV natural history
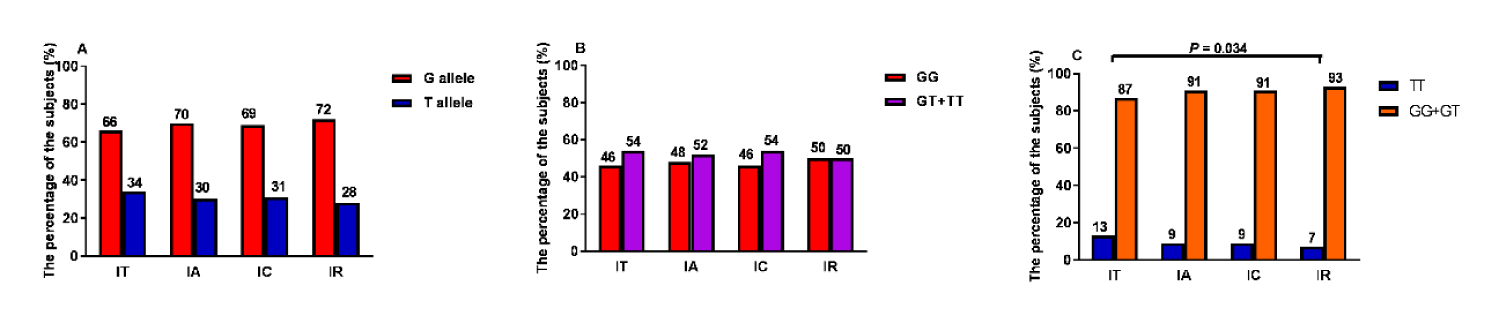
The relationship between the rs7574865 variation and HBV natural history was also analyzed. In the recession model, the frequency of rs7574865 TT genotype was higher in the IR group than the IT group (93% vs. 87%, p = 0.034; figure 3C). However, the trend was not similar in the dominant model and genotype allele between the IR and IT groups (Supplementary table 2). There was no statistical genotype significance between the IT, IA, IC, and IR groups (Supplementary table 2).
| Table 2: Predictors for HBsAg Spontaneous clearance. | ||||
| Univariate Analysis | Multivariate Analysis | |||
| HR (95% Confidence Interval) | p Value | HR (95% Confidence Interval) | p Value | |
| Gender | 1.6895 (1.4442 - 1.9764) | <0.001 | 1.3318 (1.0487 - 1.6913) | 0.019 |
| Age | 1.0695 (1.0613 - 1.0778) | <0.001 | 1.0558 (1.0455 - 1.0662) | <0.001 |
| ALT | 1.0586 (1.0510 - 1.0663) | <0.001 | 1.0492 (1.0378 - 1.0608) | <0.001 |
| AST | 1.1118 (1.0970 - 1.1267) | <0.001 | 1.0025 (0.9835 - 1.0218) | 0.800 |
| TBil | 1.0450 (1.0347 - 1.0556) | <0.001 | 1.0381 (1.0229 - 1.0535) | <0.001 |
| ALB | 0.8301 (0.8073 - 0.8535) | <0.001 | 0.8035 (0.7688 - 0.8398) | <0.001 |
| WBC | 0.8267 (0.7907 - 0.8640) | <0.001 | 0.9942 (0.9809 - 1.0077) | 0.397 |
| PLT | 0.9848 (0.9833 - 0.9863) | <0.001 | 0.9883 (0.9866 - 0.9902) | <0.001 |
| rs7574865 polymorphism | 0.8362 (0.7164 - 0.9761) | 0.023 | 0.9102 (0.7404 - 0.9235) | 0.038 |
| ALT: Alanine Aminotransferase; AST: Aspartate Aminotransferase; Tbil: Total Bilirubin; ALB: Albumin; WBC: White Blood Cell Count; PLT: Platelet Count; HR: Hazard Ratio. | ||||
Predictors associated with HBsAg spontaneous clearance
The univariate and multivariate predictors, and OR for HBsAg spontaneous clearance are shown in table 2. According to the multivariate analysis, the independent predictors for HBsAg spontaneous clearance were STAT4 rs7574865 genotype, gender, age, ALT, total bilirubin, albumin, and PLT.
HBV persistence or clearance during viral infection is dictated by the host’s complex immune response [21]. The relationship between the SNP of STAT4 and HBV clearance is a heated topic [17-19], Our study revealed a significant association between HBsAg spontaneous clearance and variations in STAT4. The prevalence of the minor allele rs7574865 T was higher in the subjects who naturally cleared HBV infection than in the CHB patients, which indicated that the subjects with rs7574865 T have a better chance of clearing the HBV infection spontaneously. However, there was no statistical significance in genotype between HBsAg clearance after treatment and STAT4 variations, nor between the IT, IA, IC, and IR groups.
STAT4 is an important cytosolic factor, which is involved in transmitting signals stimulated by I type IFN and IL12, which induce INF-γ and activate the immune response [22-24]. Recently, several studies have indicated that the polymorphisms of STAT4 contribute to autoimmune diseases [25-27]. However, few studies have identified SNP rs7574865 in STAT4 as the genetic susceptibility locus for HBV spontaneous clearance, and results were inconsistent. Lu, et al. [28] found the association between SNP rs7574865 and HBV spontaneous clearance in a study including 288 subjects. However, in two studies, Liao, et al. [29,30] failed to find the relationship between SNP rs7574865 in STAT4 and HBV spontaneous clearance. The difference in the results of these studies may be attributed to the small sample size, as well as the different regions and races. In our large sample size study, which included 3,922 subjects, we confirmed that HBV spontaneous clearance is significantly associated with the rs7574865 variation in STAT4, regardless of the allele or dominant (recessive) model analysis. Furthermore, logistic regression analysis was used to eliminate confounders, and the results were still significant. Therefore, our study indicated that variations in STAT4 play a critical role in HBV spontaneous clearance.
Nowadays, the precise mechanism by which variations in STAT4 influence natural clearance of HBV is not clear. SNP rs7574865 is located on intron 3 of STAT4 and is a non-coding region. However, rs7574865 T was reported to express more STAT4 and proteins than rs7574865 G, [17,31-32] which produced more INF-γ. A study indicated that T cells from SLE patients carrying the STAT4 risk allele T displayed an augmented response to IL-12 and induced INF-γ production [18]. INF-γ is a critical cytosolic factor to control antiviral and immune response [33]. Therefore, we speculated that subjects infected with HBV with rs7574865 T may express more STAT4 genes and proteins, produce more INF-γ, and have a higher chance of clearing HBV naturally. However, there are more factors influencing HBsAg clearance after treatment, including drug types, duration of the treatment and the host factor [18,19]. Therefore, there is no association between HBsAg clearance after treatment and STAT4 variations.
Furthermore, the relationship between CHB patients in different natural history phases and SNP rs7574865 was analyzed. The results indicated that there was no significant association between genetic variations in STAT4 and CHB natural history. In addition, genetic variations in STAT4 were not correlated with the ALT, load of HBV DNA, and HBeAg positive in CHB patients, which is consistent with the results reported by Lu, et al. [28].
Although the sample size of the study is large, there are still limitations. This is a case control study, which has a weaker evidence power than the prospective study due to lack of follow-up information. Moreover, there were more confounders than in the prospective study, although logistic regression analysis was used. Therefore, matched case control studies are warranted to obtain more potent evidence.
In summary, SNP rs7574865 in STAT4 is correlated with HBV spontaneous clearance. Subjects with the T allele had a higher chance of clearing HBV naturally than patients with the G allele. However, there is no relationship between genetic variations in STAT4 and CHB natural history phases.
We thank Dr. Haiyan Lv (Fudan University) for her helpful suggestions during the study.
This study was supported by the National Natural Science Foundation of China (81670528, 81672009, and 81871640), Shanghai municipal Health commission (NO.201740206), Shanghai Science and Technology Committee (No.18411966500) and the Joint Research Program for Emerging Frontier Technology in the Municipal Hospital of China (No. SHDC12017125).
- Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015 Oct 17;386(10003):1546-55. doi: 10.1016/S0140-6736(15)61412-X. Epub 2015 Jul 28. PMID: 26231459.
- WHO. Global hepatitis report 2017. Geneva: World Health Organization, 2017.
- Liu J, Liang W, Jing W, Liu M. Countdown to 2030: eliminating hepatitis B disease, China. Bull World Health Organ. 2019 Mar 1;97(3):230-238. doi: 10.2471/BLT.18.219469. Epub 2019 Jan 28. PMID: 30992636; PMCID: PMC6453311.
- Te HS, Jensen DM. Epidemiology of hepatitis B and C viruses: a global overview. Clin Liver Dis. 2010 Feb;14(1):1-21, vii. doi: 10.1016/j.cld.2009.11.009. PMID: 20123436.
- Guidotti LG, Isogawa M, Chisari FV. Host-virus interactions in hepatitis B virus infection. Curr Opin Immunol. 2015 Oct;36:61-6. doi: 10.1016/j.coi.2015.06.016. Epub 2015 Jul 15. PMID: 26186123; PMCID: PMC4593767.
- Matsuura K, Isogawa M, Tanaka Y. Host genetic variants influencing the clinical course of hepatitis B virus infection. J Med Virol. 2016 Mar;88(3):371-9. doi: 10.1002/jmv.24350. Epub 2015 Aug 14. PMID: 26255971.
- Martin MP, Carrington M. Immunogenetics of viral infections. Curr Opin Immunol. 2005 Oct;17(5):510-6. doi: 10.1016/j.coi.2005.07.012. PMID: 16084708.
- Belli LS, Zavaglia C, Alberti AB, Poli F, Rondinara G, Silini E, Taioli E, de Carlis L, Scalamogna M, Forti D, Pinzello G, Idèo G. Influence of immunogenetic background on the outcome of recurrent hepatitis C after liver transplantation. Hepatology. 2000 Jun;31(6):1345-50. doi: 10.1053/jhep.2000.7879. PMID: 10827162.
- Jacobson NG, Szabo SJ, Weber-Nordt RM, Zhong Z, Schreiber RD, Darnell JE Jr, Murphy KM. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J Exp Med. 1995 May 1;181(5):1755-62. doi: 10.1084/jem.181.5.1755. PMID: 7722452; PMCID: PMC2191986.
- El-Lebedy D, Raslan H, Ibrahim A, Ashmawy I, El-Aziz SA, Mohammed AM. Association of STAT4 rs7574865 and PTPN22 rs2476601 polymorphisms with rheumatoid arthritis and non-systemically reacting antibodies in Egyptian patients. Clin Rheumatol. 2017 Sep;36(9):1981-1987. doi: 10.1007/s10067-017-3632-7. Epub 2017 Apr 19. PMID: 28424905.
- Conigliaro P, Ciccacci C, Politi C, Triggianese P, Rufini S, Kroegler B, Perricone C, Latini A, Novelli G, Borgiani P, Perricone R. Polymorphisms in STAT4, PTPN2, PSORS1C1 and TRAF3IP2 Genes Are Associated with the Response to TNF Inhibitors in Patients with Rheumatoid Arthritis. PLoS One. 2017 Jan 20;12(1):e0169956. doi: 10.1371/journal.pone.0169956. PMID: 28107378; PMCID: PMC5249113.
- Ciccacci C, Conigliaro P, Perricone C, Rufini S, Triggianese P, Politi C, Novelli G, Perricone R, Borgiani P. Polymorphisms in STAT-4, IL-10, PSORS1C1, PTPN2 and MIR146A genes are associated differently with prognostic factors in Italian patients affected by rheumatoid arthritis. Clin Exp Immunol. 2016 Nov;186(2):157-163. doi: 10.1111/cei.12831. Epub 2016 Aug 2. PMID: 27342690; PMCID: PMC5054570.
- Durán-Avelar MJ, Vibanco-Pérez N, Hernández-Pacheco RR, Castro-Zambrano AD, Ortiz-Martínez L, Zambrano-Zaragoza JF. STAT4 rs7574865 G/T polymorphism is associated with rheumatoid arthritis and disease activity, but not with anti-CCP antibody levels in a Mexican population. Clin Rheumatol. 2016 Dec;35(12):2909-2914. doi: 10.1007/s10067-016-3320-z. Epub 2016 May 28. PMID: 27234231.
- Hagberg N, Joelsson M, Leonard D, Reid S, Eloranta ML, Mo J, Nilsson MK, Syvänen AC, Bryceson YT, Rönnblom L. The STAT4 SLE risk allele rs7574865[T] is associated with increased IL-12-induced IFN-γ production in T cells from patients with SLE. Ann Rheum Dis. 2018 Jul;77(7):1070-1077. doi: 10.1136/annrheumdis-2017-212794. Epub 2018 Feb 23. PMID: 29475858; PMCID: PMC6029643.
- Bi C, Li B, Cheng Z, Hu Y, Fang Z, Zhai A. Association study of STAT4 polymorphisms and type 1 diabetes in Northeastern Chinese Han population. Tissue Antigens. 2013 Mar;81(3):137-40. doi: 10.1111/tan.12057. Epub 2013 Jan 30. PMID: 23360093.
- Rueda B, Broen J, Simeon C, Hesselstrand R, Diaz B, Suárez H, Ortego-Centeno N, Riemekasten G, Fonollosa V, Vonk MC, van den Hoogen FH, Sanchez-Román J, Aguirre-Zamorano MA, García-Portales R, Pros A, Camps MT, Gonzalez-Gay MA, Coenen MJ, Airo P, Beretta L, Scorza R, van Laar J, Gonzalez-Escribano MF, Nelson JL, Radstake TR, Martin J. The STAT4 gene influences the genetic predisposition to systemic sclerosis phenotype. Hum Mol Genet. 2009 Jun 1;18(11):2071-7. doi: 10.1093/hmg/ddp119. Epub 2009 Mar 13. PMID: 19286670.
- Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, Ren WH, Long XD, Zhang H, Ma XP, Wang Z, Jiang W, Chen TY, Gao Y, Sun LD, Long JR, Huang HX, Wang D, Yu H, Zhang P, Tang LS, Peng B, Cai H, Liu TT, Zhou P, Liu F, Lin X, Tao S, Wan B, Sai-Yin HX, Qin LX, Yin J, Liu L, Wu C, Pei Y, Zhou YF, Zhai Y, Lu PX, Tan A, Zuo XB, Fan J, Chang J, Gu X, Wang NJ, Li Y, Liu YK, Zhai K, Zhang H, Hu Z, Liu J, Yi Q, Xiang Y, Shi R, Ding Q, Zheng W, Shu XO, Mo Z, Shugart YY, Zhang XJ, Zhou G, Shen H, Zheng SL, Xu J, Yu L. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2013 Jan;45(1):72-5. doi: 10.1038/ng.2483. Epub 2012 Dec 16. PMID: 23242368; PMCID: PMC4105840.
- Jiang DK, Wu X, Qian J, Ma XP, Yang J, Li Z, Wang R, Sun L, Liu F, Zhang P, Zhu X, Wu J, Chen K, Conran C, Zheng SL, Lu D, Yu L, Liu Y, Xu J. Genetic variation in STAT4 predicts response to interferon-α therapy for hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2016 Apr;63(4):1102-11. doi: 10.1002/hep.28423. Epub 2016 Feb 19. PMID: 26704347.
- Chen H, Sun J, Zhou B, Xie Q, Liang X, Fan R, Conran C, Xu J, Ji Y, Zhang X, Sun L, Jia J, Wang G, Hou J, Jiang DK. Variants in STAT4 Associated With Cure of Chronic HBV Infection in HBeAg-positive Patients Treated With Pegylated Interferon-alpha. Clin Gastroenterol Hepatol. 2020 Jan;18(1):196-204.e8. doi: 10.1016/j.cgh.2019.04.044. Epub 2019 Apr 28. PMID: 31042581.
- Terrault NA, Bzowej NH, Chang KM, Hwang JP, Jonas MM, Murad MH; American Association for the Study of Liver Diseases. AASLD guidelines for treatment of chronic hepatitis B. Hepatology. 2016 Jan;63(1):261-83. doi: 10.1002/hep.28156. Epub 2015 Nov 13. PMID: 26566064; PMCID: PMC5987259.
- Rehermann B, Fowler P, Sidney J, Person J, Redeker A, Brown M, Moss B, Sette A, Chisari FV. The cytotoxic T lymphocyte response to multiple hepatitis B virus polymerase epitopes during and after acute viral hepatitis. J Exp Med. 1995 Mar 1;181(3):1047-58. doi: 10.1084/jem.181.3.1047. PMID: 7532675; PMCID: PMC2191941.
- Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DA, Doherty PC, Grosveld GC, Ihle JN. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996 Jul 11;382(6587):171-4. doi: 10.1038/382171a0. PMID: 8700208.
- Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O’Shea JJ, Biron CA. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science. 2002 Sep 20;297(5589):2063-6. doi: 10.1126/science.1074900. PMID: 12242445.
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009 Nov;9(11):798-809. doi: 10.1038/nrc2734. PMID: 19851315; PMCID: PMC4856025.
- Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, de Bakker PI, Le JM, Lee HS, Batliwalla F, Li W, Masters SL, Booty MG, Carulli JP, Padyukov L, Alfredsson L, Klareskog L, Chen WV, Amos CI, Criswell LA, Seldin MF, Kastner DL, Gregersen PK. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007 Sep 6;357(10):977-86. doi: 10.1056/NEJMoa073003. PMID: 17804842; PMCID: PMC2630215.
- Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, Guiducci C, Chen R, Alfredsson L, Amos CI, Ardlie KG; BIRAC Consortium, Barton A, Bowes J, Brouwer E, Burtt NP, Catanese JJ, Coblyn J, Coenen MJ, Costenbader KH, Criswell LA, Crusius JB, Cui J, de Bakker PI, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Lee AT, Liu X, Martin P, Morgan AW, Padyukov L, Posthumus MD, Radstake TR, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van Mil AH, van der Horst-Bruinsma IE, van der Schoot CE, van Riel PL, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth BP; YEAR Consortium, Wijmenga C, Karlson EW, Toes RE, de Vries N, Begovich AB, Worthington J, Siminovitch KA, Gregersen PK, Klareskog L, Plenge RM. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010 Jun;42(6):508-14. doi: 10.1038/ng.582. Epub 2010 May 9. PMID: 20453842; PMCID: PMC4243840.
- Korman BD, Kastner DL, Gregersen PK, Remmers EF. STAT4: genetics, mechanisms, and implications for autoimmunity. Curr Allergy Asthma Rep. 2008 Sep;8(5):398-403. doi: 10.1007/s11882-008-0077-8. PMID: 18682104; PMCID: PMC2562257.
- Lu Y, Zhu Y, Peng J, Wang X, Wang F, Sun Z. STAT4 genetic polymorphisms association with spontaneous clearance of hepatitis B virus infection. Immunol Res. 2015 Jun;62(2):146-52. doi: 10.1007/s12026-015-8645-1. PMID: 25829184.
- Liao Y, Cai B, Li Y, Chen J, Ying B, Tao C, Zhao M, Ba Z, Zhang Z, Wang L. Association of HLA-DP/DQ, STAT4 and IL-28B variants with HBV viral clearance in Tibetans and Uygurs in China. Liver Int. 2015 Mar;35(3):886-96. doi: 10.1111/liv.12643. Epub 2014 Aug 5. PMID: 25041342.
- Liao Y, Cai B, Li Y, Chen J, Ying B, Tao C, Zhao M, Ba Z, Zhang Z, Wang L. Association of HLA-DP/DQ, STAT4 and IL-28B variants with HBV viral clearance in Tibetans and Uygurs in China. Liver Int. 2015 Mar;35(3):886-96. doi: 10.1111/liv.12643. Epub 2014 Aug 5. PMID: 25041342.
- Lamana A, López-Santalla M, Castillo-González R, Ortiz AM, Martín J, García-Vicuña R, González-Álvaro I. The Minor Allele of rs7574865 in the STAT4 Gene Is Associated with Increased mRNA and Protein Expression. PLoS One. 2015 Nov 16;10(11):e0142683. doi: 10.1371/journal.pone.0142683. PMID: 26569609; PMCID: PMC4646635.
- Abelson AK, Delgado-Vega AM, Kozyrev SV, Sánchez E, Velázquez-Cruz R, Eriksson N, Wojcik J, Linga Reddy MV, Lima G, D’Alfonso S, Migliaresi S, Baca V, Orozco L, Witte T, Ortego-Centeno N; AADEA group, Abderrahim H, Pons-Estel BA, Gutiérrez C, Suárez A, González-Escribano MF, Martin J, Alarcón-Riquelme ME. STAT4 associates with systemic lupus erythematosus through two independent effects that correlate with gene expression and act additively with IRF5 to increase risk. Ann Rheum Dis. 2009 Nov;68(11):1746-53. doi: 10.1136/ard.2008.097642. Epub 2008 Nov 19. PMID: 19019891; PMCID: PMC3878433.
- Morinobu A, Gadina M, Strober W, Visconti R, Fornace A, Montagna C, Feldman GM, Nishikomori R, O’Shea JJ. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc Natl Acad Sci U S A. 2002 Sep 17;99(19):12281-6. doi: 10.1073/pnas.182618999. Epub 2002 Sep 4. PMID: 12213961; PMCID: PMC129436.
| Supplementary table 1: Impact of STAT4 rs7574865 polymorphism on HBV outcome. | ||||||||||
| a | b | c | d | p value | ||||||
| HC (n = 1,012) | SC (n = 1,052) | CHB (n = 1,688) | TC (n = 170) | a vs. b | a vs. c | a vs. d | b vs. c | b vs. d | c vs. d | |
| STAT4 polymorphism | ||||||||||
| GG | 474 (47%) | 464 (44%) | 819 (49%) | 90 (53%) | 0.269 | 0.103 | 0.272 | 0.056 | 0.098 | 0.538 |
| GT | 426 (42%) | 480 (46%) | 724 (43%) | 66 (39%) | ||||||
| TT | 112 (11%) | 108 (10%) | 145 (9%) | 14 (8%) | ||||||
| Recessive model | ||||||||||
| GG+GT | 900 (89%) | 944 (90%) | 1,543 (91%) | 156 (92%) | 0.556 | 0.034 | 0.268 | 0.141 | 0.412 | 0.875 |
| TT | 112 (11%) | 108 (10%) | 145 (9%) | 14 (8%) | ||||||
| Dominant model | ||||||||||
| GG | 474 (47%) | 464 (44%) | 819 (49%) | 90 (53%) | 0.213 | 0.397 | 0.140 | 0.023 | 0.032 | 0.272 |
| GT+TT | 538 (53%) | 588 (56%) | 869 (51%) | 80 (47%) | ||||||
| Allele | ||||||||||
| G | 1,374 (68%) | 1,408 (67%) | 2,362 (70%) | 246 (72%) | 0.508 | 0.109 | 0.101 | 0.018 | 0.047 | 0.359 |
| T | 650 (32%) | 696 (33%) | 1,014 (30%) | 94 (28%) | ||||||
| Values are expressed as number of subjects (%). HC: HBV Uninfected; SC: Hepatitis B Surface Antigen (Hbsag) Spontaneous Clearance; CHB: Chronic Hepatitis B; TC: Hbsag Clearance after Treatment. |
||||||||||
| Supplementary table 2: Impact of STAT4 rs7574865 polymorphism on chronic hepatitis B patients. | ||||||||||
| a | b | c | d | p value | ||||||
| IT ( n = 97) | IA (n = 855) | IC (n = 198) | IR (n = 538) | a vs. b | a vs. c | a vs. d | b vs. c | b vs. d | c vs. d | |
| STAT4 polymorphism | ||||||||||
| GG | 45 (46%) | 412 (48%) | 92 (46%) | 270 (50%) | 0.349 | 0.491 | 0.107 | 0.908 | 0.446 | 0.526 |
| GT | 39 (40%) | 367 (43%) | 88(45%) | 230 (43%) | ||||||
| TT | 13 (14%) | 76 (9%) | 18 (9%) | 38 (7%) | ||||||
| Recessive model | ||||||||||
| GG+GT | 84 (87%) | 779 (91%) | 180 (91%) | 500 (93%) | 0.148 | 0.257 | 0.034 | 0.928 | 0.226 | 0.358 |
| TT | 13 (13%) | 76 (9%) | 18 (9%) | 38 (7%) | ||||||
| Dominant model | ||||||||||
| GG | 45 (46%) | 412 (48%) | 92 (46%) | 270 (50%) | 0.737 | 0.991 | 0.491 | 0.662 | 0.468 | 0.371 |
| GT+TT | 52 (54%) | 443 (52%) | 106 (54%) | 268 (50%) | ||||||
| Allele | ||||||||||
| G | 129 (66%) | 1,191(70%) | 272 (69%) | 770 (72%) | 0.367 | 0.592 | 0.153 | 0.708 | 0.282 | 0.282 |
| T | 65 (34%) | 519 (30%) | 124 (31%) | 306 (28%) | ||||||
| Values are expressed as number of subjects (%). IT: Immune-Tolerant Phase; IA: Immune-Tolerant Phase; IC: Inactive Chronic Hepatitis B Phase; IR: Immune Reactivation Phase. |
||||||||||
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.