> Biology. 2020 October 09;1(6):201-212. doi: 10.37871/jbres1144.
-
Subject area(s):
- Infectious Diseases
- Antivirology
- Antiretrovirology
- Biology
Gossypetin Derivatives are also Putative Inhibitors of SARS-COV 2: Results of a Computational Study
Anna-Gaelle Giguet-Valard1,2*, Kevin Raguette1, Stephanie Morin1, Remi Bellance2 and Juliette Smith Ravin1
2University Hospital Center of Martinique
- 3CL Protease
- SARS-COV2
- Gossypetin-3'-O-glucoside
- Molecular docking
- Free binding energy
- Natural polyphenols compounds
SARS-CoV-2 is the third most highly virulent human coronavirus of the 21st century. It is linked with fatal respiratory illness. Currently, there are still no effective treatments of Covid-19. Among many drugs evaluated, few have proven conclusive clinical efficacy. Furthermore, the spread of the disease mandates that ideal medications against Covid-19 be cheap and available worldwide. Therefore, there is a rationale to evaluate whether treatments of natural origin from aromatic and medicinal plants have the ability to prevent and/or treat COVID-19. We evaluated in this study the inhibition of COVID-19 protease by natural plants compounds such as Gossypetin-3'-O-glucoside (G3'G). G3'G has been isolated from the petals of Talipariti elatum Sw. found almost exclusively in Martinique. It has no crystallography or modelisation studies. Antifungal and antioxidant properties are already published. We study its binding affinity so potential inhibition capability against SARS-CoV2 3CLpro mean protease as compared to other previously tested natural or pharmacological molecules by molecular docking. We propose Gossypetin derivatives as good tropical natural compounds candidate that should be further investigated to prevent or treat COVID19.
Severe Acute Respiratory Syndrome (SARS) was the first new infectious disease identified in the twenty-first century. This acute and often severe, respiratory illness is caused by a new coronavirus, SARS-Coronavirus (SARS- CoV). Between 2002-2003, the first outbreak of SARS reached all five continents by air-travel routes.
SARS-CoV is a zoonotic virus that resides in hosts that form its natural reservoir, such as bats. It can also infect intermediate hosts, such as small animals (for example, palm civets), before being transmitted to humans. SARS-CoV can infect and replicate in several cell types in humans and cause serious organ injuries [1].
A novel coronavirus strain linked with fatal respiratory illness was reported in late 2019. At the beginning of 2020, the World Health Organization (WHO) permanently named the 2019-nCoV pathogen as SARS-CoV-2 and the causing disease as Coronavirus Disease 2019 (COVID-2019). SARS-CoV-2 is the third most highly virulent human coronavirus of the 21st century followed by the SARS-COV and MERS-COV. SARS-COV-2 is the seventh strain of human coronavirus. It is taxonomically placed in the Genre Beta coronavirus, and exhibits 89.1% and 60% nucleotide sequence similarity with SARS and MERS coronaviruses, respectively [2,3].
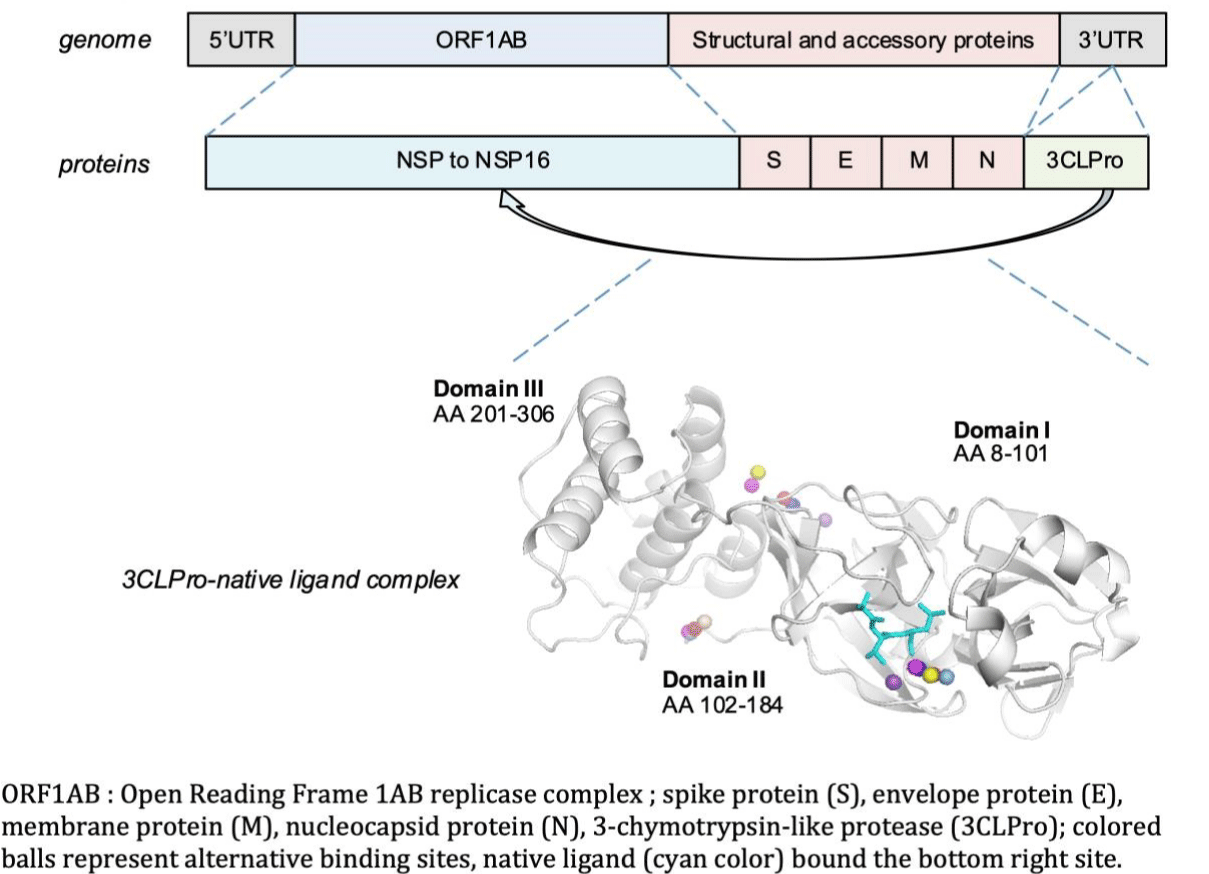
Coronaviruses are large enveloped positive single strand RNA viruses. They are highly evolving, with a high frequency of gene recombination and mutation [4,5]. Like other beta-coronaviruses, the genomic organization of SARS-CoV-2 consists in a 5'-Untranslated Region (UTR), a replicase complex ORF1AB encoding Non-Structural Proteins (NSPS), a Spike Protein (S) gene, an Envelope Protein (E) gene, a Membrane Protein (M) gene, a Nucleocapsid Protein (N) gene, and a 3'-UTR containing several unidentified non-structural open reading frames (Figure 1).
Usually, beta-coronaviruses genome transcribes an approximately 800 kDa polypeptide. This polypeptide is proteolytically cleaved to generate various proteins. The proteolytic process is mediated by a Papain-Like Protease (PLpro) and a 3-Chymotrypsin-Like Protease (3CLpro). 3CLpro gene is located at the 3'-end and exhibits a high-degree variability [5]. The 3CLpro cleaves the polyprotein at 11 distinct sites, generating various non-structural proteins involved in the virus replication. The functional importance of 3CLpro makes it an attractive target for the development of effective antiviral drugs [6,7]. 3CLpro is considered as a validated potential drug non-toxic target for anti-coronaviruses inhibitors screening [2]. Screening and repurposing of Food and Drug Administration approved antiviral drugs to inhibit 3CLpro may provide an alternative fast-track approach for identifying and developing new treatments for SARS-COV-2 infection [5].
Like other coronaviruses, the Three-Dimensional (3-D) structure of SARS-Cov-2 3CLpro consists of three domains. Domain I (8-101 amino acid residues) and II (102-184 amino acid residues) are essentially beta-barrels, and bear a resemblance to chymotrypsin. Domain III (201-306 amino acid residues) is mainly comprised of alpha-helices. The substrate binding region is located at the cleft of Domain I and II. In addition to the catalytic center, amino acid residues of subsites designated as S1 to S5, play a key role in natural substrate binding: Thr25, His163, Glu166, Cys145, Gly143, His172, Phe140, Met49, His41, Met165, Asn142 and Gln189. His41 and Cys145 3CLpro residues are considered to be critical for the virus activity. 3CLpro/ligand dyad is crucial for the catalytic activity and essential in viral replication [5].
Currently, there are still no effective treatments of Covid-19. Development of new treatments may require months to years. So far, clinical trials have been quite disappointing, and drug toxicity issues have been raised, notably for chloroquine [8,9]. Therefore, there is a rationale to evaluate whether treatments of natural origin from aromatic and medicinal plants have the ability to prevent and/or treat COVID-19 [4,10,11].
A joint research team of the Shanghai Institute of Materia Medica and Shanghai Tech University performed drug screening in silico, and enzyme activity test.
They identified thus far 30 agents with potential efficacy against COVID-19, including Western medicines, natural products, and traditional Chinese medicines. They also found that Chinese herbal medicines such as Rhizoma Polygoni Cuspidati and Radix Sophorae Tonkinensis may contain active ingredients against SARS-COV-2 [12].
Computational prediction by molecular docking is a reliable method to study interactions between candidate molecules and proteins. It is widely used in sillico experiments to screen drugs inhibition efficacy. Molecular docking is a kind of bioinformatics modelling which involves the interaction of two or more molecules to give the stable adduct. Docking involves the placement of a ligand within a binding site and the prediction of the free energy of binding for such poses. In molecular docking, the most important aspect is the calculation of binding energy so as to fit a ligand in a binding site. It is actually a reference method used to find agent against nCoV-2019 [13].
Very few data is available on the clinical use of medicinal plants to prevent or cure COVID-19. However, scientists' teams worldwide (India, China, Iran, Morocco, sub-Saharian African) have published computer-based prediction studies of molecules from their own pharmacopoeia, mostly targeting the binding affinity of the main SARS-CoV-2 3CLprotease [5,14-16]. Therefore, we evaluated in this study the inhibition of COVID-19 protease by herbal plants compound found in Martinique.
Martinique is a tropical Caribbean island enjoying a large biodiversity. The population has a large use of local medicinal plants. Some proteins from Caribbean medicinal plants have already been identified in protein database for molecular docking tests. Many others have not, such as Gossypetin-3'-O-glucoside (G3'G). This molecule, isolated from the petals of Talipariti elatum Sw. Found, almost exclusively in Martinique. It has no crystallography modelisation studies, despite reported antifungal and antioxidant properties [17].
In this study we searched to model G3'G, and study its binding affinity so potential inhibition capability against SARS-CoV2 3CLpro mean protease as compared to other previously tested natural or pharmacological molecules.
Molecules
We selected pharmaceutical molecules included in the Guidelines (version 6) for treatment of COVID19 and molecules from medicinal plants. We defined a list of molecules allowing us to conclude about the effectiveness of our method and classify new candidate molecules. It is the reason why we also selected compounds that have already been computationally tested. We tested some drugs and medicinal plants molecules for the first time by molecular docking (Table 1).
Table 1: Dataset of molecules. Shows PubChem Id, chemical formula of molecules of interest, usual trade name of drugs and plants species origin. P means medicinal plants origin and D means drugs. |
||||||
| Name | Type | Class | Trade name/species | PubChem Id | Chemical formula | Reference |
| Aloenin | P | Anthraquinones | Aloe vera | CID 162305 | C19H22O10 | [1] |
| Amiodarone | D | Class III Antiarrhythmic | Cordarone | CID 2157 | C25H29I2NO3 | [1] |
| Arbidol | D | Antiviral | Umifénovir | CID 131411 | C22H25BrN2O3S | [2] |
| Astragalin | P | Flavonol | Harsingar | CID 5282102 | C21H20O11 | [3] |
| Beta-Eudesmol | P | Anti-Angiogenic/Anti-Tumor, Sesquiterpenoid | Laurel (lauris nobilis L) | CID 91457 | C15H26O | [4] |
| Cannabidiol | P | Cannabinoid | Cannabis | CID 644019 | C21H30O2 |
|
| Chloroquine | D | Antimalarial Of The 4-Aminoquinoline Family | Axemal, Dolquine, Quensyl, Plaquenil | CID 2719 | C18H26ClN3 | [5] |
| Crocin | P | Carotenoid | saffron (crocus sativus) | CID 5281233 | C44H64O24 | [4] |
| Curcumin | P | Polyphenol | Turmeric | CID 969516 | C21H20O6 | [1] |
| Digitoxigenin | P | Cardenolide | laurel (nerium oleander) | CID 4369270 | C23H34O4 | [4] |
| Gossypetin-3-Glucoside | P | Flavonoid | Hibiscus (Hibiscus sabdariffa L/tiliaceus) | CID 44259979 | C21H20O13 | [6] |
| Gossypetin-3'-O-Glucoside | P | Flavonoid | Hibiscus (Abelmoschus manihot/Hibiscus elatus Sw) | CID 99649 | C21H20O13 | [6] |
| Gossypetin-7-O-Glucoside | P | Flavonoid | Hibiscus (sabdariffa L./tiliaceus/elatus Sw.) | CID 90658329 | C21H19O13- | [6] |
| Gossypetin-8-Glucoside | P | Flavonoid | Hibiscus (vitifolius/sabdariffa L) | CID 5281621 | C21H20O13 | [6] |
| Hydroxychloroquine | D | Antimalarial Of The 4-Aminoquinoline Family | Axemal, Dolquine, Quensyl, Plaquenil | CID 3652 | C18H26ClN3O |
|
| Kaempferol | P | Flavonol | spinach, cabbage, dill, chinese cabbage, katuk | CID 5280863 | C15H10O6 | [1] |
| Lopinavir | D | Antiviral | Kaletra (ritonavir + lopinavir) | CID 92727 | C37H48N4O5 |
|
| Luteolin-7-Glucoside | P | Flavones | olive, star fruit, chili pepper, welsh onion, leek | CID 5280637 | C21H20O11 | [1,5] |
| Native Ligand |
|
|
|
|
C35H53N6O8 | [7] |
| Nelfinavir | D | Antiretroviral | Viracept | CID 64143 | C32H45N3O4S | [8] |
| Nictoflorin | P | Flavonoid | Harsingar | CID 5318767 | C27H30O15 | [1] |
| Piceid | P | Polyphenol | Picea sitchensis/ Reynoutria japonica | CID 5281718 | C20H22O8 | [9] |
| Quercetin | P | Flavonoid | dill, fennel leaves, onion, oregano, chili peppe | CID 5280459 | C15H10O7 | [3] |
| Quercitrin | P | Flavonoid | Wheat (Fagopyrum tataricum)/Oak (Quercus alba/robur) | CID 5280459 | C21H20O11 | [6] |
| Remdesivir | D | Antiviral | Remdesivir | CID 121304016 | C27H35N6O8P | [5] |
| Ribavirin | D | Antiviral | Ibavyr | CID 37542 | C8H12N4O5 | [2,5] |
Three-Dimensional (3D) dataset
3CLpro and ligands: The majority of biological macromolecules crystal structures is collected in PDB worldwide archive [18].
SARS-CoV-2 3-chymotrypsin-like protease (3CLpro) (PDB ID: 6LU7) structures were obtained from PDB (https://www.rcsb.org/). The native ligand for 6LU7 is n-[(5-methylisoxazol-3-yl) carbonyl] alanyl-l-valyl-n~1~-((1r,2z)-4-(benzyloxy)-4-oxo-1-{[(3r)-2-oxopyrrolidin-3-yl]methyl}but-2-eny-l-leucinamide [19,20].
Other three-dimensional structures of molecules of interest were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/). It is a chemical substance and biological activities repository consisting of three databases, including substance, compound, and bioassay databases [21].
3-D structures of the following molecules of interest were obtained in pdf format:
- Natural molecules: Kaempferol (CID 5280863), Quercetin (CID 5280459), Luteolin- 7-Glucoside (CID 5280637), Aloenin (CID 162305), Cannabidiol (CID 644019), Curcumin (CID 969516), Nictoflorin (CID 5318767), Astragalin (CID 5282102), Digitoxigenin (CID 4369270), β-eudesmol (CID 91457), Gossypetin-8-glucoside (CID 5281621), Gossypetin-7-O-glucoside (CID 90658329), Gossypetin-3-glucoside (CID 44259979), Piceid (CID 5281718), Emodin-8-glucoside (CID 99649), Crocin (CID 5281233), and Gossypetin3'-O-glucoside.
- Pharmaceutical drugs: Nelfinavir (CID 64143), Hydroxychloroquine (CID 3652), Chloroquine (CID 2719), Amiodarone (CID 2157), Remdesivir (CID 121304016), Ribavirin (CID 37542), Arbidol (CID 131411), Lopinavir (CID 92727), Quercitrin (CID 5280459).
- Modelisation: Gossypetin-3'-O-glucoside (G3'G) is not present in PDB norPubChem database. Using Opsin [22]we determined that G3'G international chemical name is 3,5,7,8-tetrahydroxy-2-(4-hydroxy-3-{[(3S,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan- 2-yl]oxy}phenyl)-4H-chromen-4-one,allowing us to convert it in SMILES. We used also the "SMILES" of Crocin, and Lopinavir, which have no 3D model proposition. We use "-h et –gen3D" options of Openbabel software to obtain a 3-D model of these 3 molecules in pdf format [23].
In silico method
Lupinski's rule: According to Lupinski's rule, efficient drugs should have good permeation and oral absorption depending on respect of the following 5 physical and chemical properties: molecular weights > 500, C logP> 5, more than 5 hydrogen-bond donors, and more than 10 acceptor groups. An orally active drug has no more than one violation of the following criteria [24]. Based on Lupinski's rules, we checked druglikeness of best candidate. We use RDKIT (open-source cheminformatics; https://www.rdkit.org) Lipinski's rule of five.
Determination of 3CLpro active sites: The detection of binding sites on the surface of the protein was done with MetaPocket 2.0 (MPT) [25]. This meta-server uses a consensual method, in which binding sites are predicted from eight methods: LIGSITEcs (LCS), PASS (PAS), Q-SiteFinder (QSF), SURFNET (SFN), Fpocket (FPK), GHECOM (GHE), ConCavity (CON) and POCASA (PCS). The results are combined to maximize the rate of successful prediction.
Molecular Docking: Pymol version 1.7.2.1 was used for protein optimization, by removing ligand, water and other atoms, and then adding a polar hydrogen group. Protonation at pH7.4 using amberff99 force field has been done with pdb2pqr version 2.1.0 and PROPKA algorithm [26,27]. Optimal structure of the protein was then saved as .pdf format. We converted all ligands in .mol2 format at pH7.4 with Openable software [23]. We generated several conformers and minimized theirs structures using MMFF94 force field. We used only the lowest energy conformers. Native ligand has not been minimized. For both ligands and protein, allocation of Gasteiger atomic partial charges and polar hydrogens were done with scripts from Auto Dock Tools Mgl tools 1.5.6: prepare_receptor4.py and prepare_ligand4.py. The docking site on protein target was defined by establishing a grid box with a default grid spacing centered on the position of site native ligand. The docking analyses were performed with Autodock 4.2.6, AutodockVina version 1.1.2, and Smina version Oct 15 2019. AutoDock uses a Lamarckian genetic algorithm and an empirical free energy force field scoring function [28]. AutoDockVina employs an iterated local search global optimizer and an hybrid scoring function (empirical + knowledge based function) [29]. Smina is a fork of AutoDockVina that uses a scoring function called Vinardo, with enhanced features based on AutoDockVina [30]. To obtain free binding energy we made several per methods. We performed fifty docking runs per ligand with Autodock 4.2.6 and Smina method. Only twenty runs with AutodockVina version 1.1.2 which cannot do more [28,31].
Consensus scoring (C-score): To evaluate docking efficacy, we measured the number of favorable intermolecular interactions such as hydrogen bonds and hydrophobic contacts. But to classify which ligands are most likely to interact strongly with the protein 3CLpro, we rank them according to a consensus scoring method [32]. First, for each method we determined a score (S) such as: S = φ - μ / σ. Where φ is the value of free binding energy of a molecule; μ is the mean value of the set of molecules; σ is the standard deviation observed for the set of molecules. Then, we normalized the score S to S', where S'= (S-Smin)/(Smax – Smin). Where, Smax and Smin, are the maximum and the minimum S-score value of the set of molecules of a method. Finally, we attribute a Consensus-Score (C-score) for each molecule. This value is the sum of the three S'-score obtained for each molecule. Our ranking is based on the sorting of molecules respecting to their consensus-score.
Binding site determination
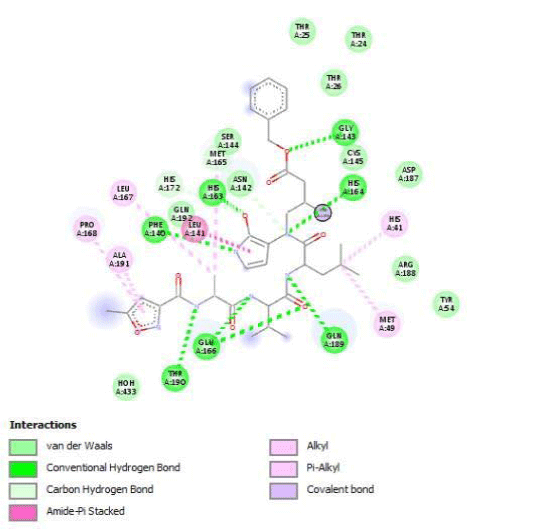
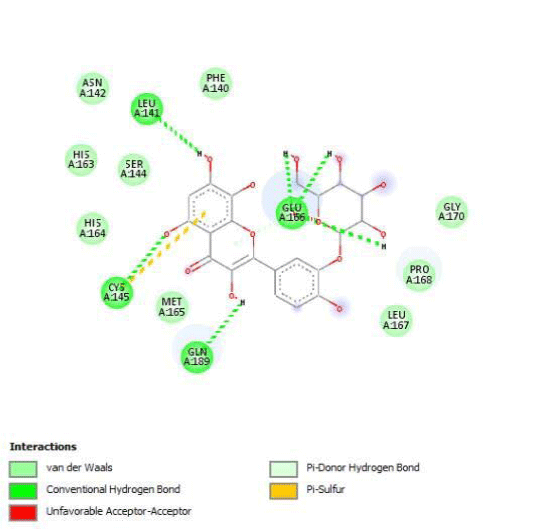
The determination of Amino Acids (AA) in the active site is used to analyze the Grid box and docking evaluation results. We identify residues of the active site by visualization of 3CLpro/native ligand linkages. We found several kinds of atomic links with critical AA of 3CL/Pro:
- H-bonds: Gly143, Phe140, His163, His164, Glu166, Gln189 and Thr190
- Amide Pi-stackedwithLeu141
- pi-alkyle bounds with His41, Met49, Leu167, Pro168 and Ala191
- Carbones-hydrogen bounds with Met165 and His172
- Van der Walls interactions on 11 AA including Asn142, Asp187, Arg188 and Tyr54.
Molecular docking analyses
Three-dimensional models of G3'G, Crocin and Lopinavir, which were not previously available, could be obtained using Openbabel software, and were subsequently used in the docking analysis.
Table 2 presents free binding energy (ΔG) scores found with each molecular docking method, and the T-value. Crocin has the longest carbon chain of the set. Free binding energy of the Crocin is not calculated by Autodock4 because it has too many twists. Autodock4 accepts a maximum number of 32 twists per molecules.
Table 2: Free binding energy (ΔG) by docking methods and consensus scores. |
|||||
| Name | Type | Autodock4 | Autodock Vina | Smina | Consensus Score |
| Aloenin | P | -7,47 | -7,2 | -8,01 | 1,476 |
| Amiodarone | D | -9,99 | -6,8 | -7 | 1,619 |
| Arbidol | D | -9,38 | -6,9 | -7,01 | 1,634 |
| Astragalin | P | -10,39 | -8,1 | -9,49 | 0,609 |
| Beta-eudesmol | P | -7,3 | -5,9 | -6,07 | 2,347 |
| Cannabidiol | P | -7,63 | -6 | -6,2 | 2,257 |
| Chloroquine | D | -7,64 | -6 | -5,88 | 2,326 |
| Crocin | P | NA | -6,8 | -7,94 | 2,251 |
| Curcumin | P | -8,57 | -6,8 | -7,06 | 1,725 |
| Digitoxigenin | P | -7,79 | -6,8 | -7,23 | 1,753 |
| Gossypetin-3-glucoside | P | -11,93 | -8 | -9,69 | 0,784 |
| Gossypetin-3'-O-glucoside | P | -11,78 | -7,5 | -9,07 | 0,47 |
| Gossypetin-7-O-glucoside | P | -10,66 | -8,2 | -9,74 | 0,499 |
| Gossypetin-8-glucoside | P | -11,62 | -7,9 | -9,66 | 0,536 |
| Hydroxychloroquine | D | -6,79 | -6,2 | -6,23 | 2,255 |
| Kaempferol | P | -8,87 | -7,7 | -8,49 | 1,088 |
| Lopinavir | D | -8,59 | -8,9 | -9,1 | 0,578 |
| Luteolin-7-glucoside | P | -10,66 | -8,4 | -9,73 | 0,434 |
| NATIVE LIGAND | -8,23 | -7,6 | -8,1 | 1,26 | |
| Nelfinavir | D | -9,44 | -8,2 | -8,66 | 0,836 |
| nictoflorin | P | -11,4 | -8,7 | -10,47 | 0,111 |
| Piceid | P | -9,12 | -7,7 | -8,78 | 1,004 |
| Quercetin | P | -9,68 | -7,3 | -8,32 | 1,19 |
| Quercitrin | P | -11,88 | -8,4 | -9,81 | 0,315 |
| Remdesivir | D | -8,03 | -8,1 | -8,55 | 1,012 |
| Ribavirin | D | -7,34 | -6 | -6,49 | 2,219 |
The consensus-score of ΔG values range from 0,111 to 2,347. G3'G is found with one of the lowest values which sign a best affinity to link 3CLPro.
We evaluated our products regarding Lupinski rules compliance and others Autodock 4 docking data (Table 3). The affinity of drug compounds is evaluated through the number and type (Hydrogen or hydrophobic type) of bonds, which occur with the active site of the protein. The lower is inhibition constant the more is affinity. While desolvation energy and intermolecular energy are proportional to affinity. Nictoflorin and Crocin present 3 violations. Chloroquine / Hydroxychloroquine and Ribavirin are the only drugs that show no violation. Among the natural compounds, Aloenin, Beta-eudesmol, Curcumin, Digitoxigenin, Kaempferol, Quercetin show no violation either. These are proteins from various classes: anthraquinones, sesquiterpenoid, polyphenol, cardenolide, flavonol, and flavonoid.
Table 3: Other docking data regards to Lupinski rules violations. |
|||||||||
|
|
Lupinski Rules | Docking DATA | ||||||
| Name | Type | Molecular weight | LogP | H-Bond donor | H-bond acceptor | Violation | Inhibition Constant | Intermolecular Energy | VDW-H Bond Desolvation Energy |
| Aloenin | P | 410,375 | -0,492 | 5 | 10 | 0 | 3,32 | -8,36 | -8,36 |
| Amiodarone | D | 645,319 | 6,936 | 0 | 4 | 2 | 47,76 | -12 | -12 |
| Arbidol | D | 477,424 | 5,177 | 1 | 6 | 1 | 132,95 | -9,4 | -9,4 |
| Astragalin | P | 448,38 | -0,245 | 7 | 11 | 2 | 24,24 | -11,05 | -11,05 |
| Beta-Eudesmol | P | 222,372 | 3,92 | 1 | 1 | 0 | 4,43 | -7,91 | -7,91 |
| Cannabidiol | P | 314,469 | 5,847 | 2 | 2 | 1 | 2,53 | -9,89 | -9,89 |
| Chloroquine | D | 319,88 | 4,811 | 1 | 3 | 0 | 2,53 | -9,86 | -9,86 |
| Crocin | P | 976,972 | -5,225 | 14 | 24 | 3 |
|
|
|
| Curcumin | P | 368,385 | 3,37 | 2 | 6 | 0 | 523,22 | -10,11 | -10,11 |
| Digitoxigenin | P | 374,521 | 3,604 | 2 | 4 | 0 | 1,96 | -8,68 | -8,68 |
| Gossypetin-3-Glucoside | P | 480,378 | -0,833 | 9 | 13 | 2 | 2,31 | -10,84 | -9,61 |
| Gossypetin-3'-O-Glucoside | P | 480,378 | -0,833 | 9 | 13 | 2 | 1,8 | -9,61 | -10,84 |
| Gossypetin-7-O-Glucoside | P | 479,37 | -1,465 | 8 | 13 | 2 | 15,38 | -8,66 | -8,66 |
| Gossypetin-8-Glucoside | P | 480,378 | -0,833 | 9 | 13 | 2 | 3,04 | -8,84 | -8,84 |
| Hydroxychloroquine | D | 335,879 | 3,783 | 2 | 4 | 0 | 10,5 | -8,48 | -8,48 |
| Kaempferol | P | 286,239 | 2,282 | 4 | 6 | 0 | 317,27 | -9,89 | -9,89 |
| Lopinavir | D | 628,814 | 4,328 | 4 | 5 | 1 | 507,05 | -10,88 | -10,88 |
| Luteolin-7-Glucoside | P | 448,38 | -0,244 | 7 | 11 | 2 | 15,22 | -10,64 | -10,64 |
| Native Ligand |
|
684,835 | 2,099 | 6 | 10 | 2 | 919,78 | -14,03 | -14,03 |
| Nelfinavir | D | 567,796 | 4,748 | 4 | 6 | 1 | 119,44 | -11,79 | -11,79 |
| Nictoflorin | P | 594,522 | -1,393 | 9 | 15 | 3 | 4,41 | -10,76 | -10,76 |
| Piceid | P | 390,388 | 0,447 | 6 | 8 | 1 | 207,24 | -10,66 | -10,66 |
| Quercetin | P | 302,238 | 1,988 | 5 | 7 | 0 | 80,71 | -9,38 | -9,38 |
| Quercitrin | P | 448,38 | 0,489 | 7 | 11 | 2 | 1,96 | -10,86 | -10,86 |
| Remdesivir | D | 602,585 | 2,312 | 4 | 13 | 2 | 1,3 | -10,81 | -10,81 |
| Ribavirin | D | 244,207 | -3,011 | 4 | 8 | 0 | 4,18 | -6,34 | -6,34 |
G3'G/ Gossypetin-3-Glucoside (G3G)/ Gossypetin-8-Glucoside (G8G), Digitoxigenin, Quercitrin, and Cannabidiol among natural products; Remdesivir, Chloroquine and Ribavirin among synthetic products, had the best inhibition constants. All Gossypetins derivatives, Nictoflorin and Crocin presented each more than 10 H-bonds with the protein. Remdesivir has 13 H-bonds acceptor and only 4 H bonds donor.
By matching ranking of the best intermolecular energy and desolvation energy ligands, we find that G3'G/G8G, Cannabidiol and Chloroquine are all time present in the top 10 ranking.
Finally, the products analysed were presented according to their origin (natural origin or chemical synthesis), and their rank in consensus score of free binding energy.
The compounds with the 6 more favourable free binding energy were natural molecules from medicinal plants (Table 4). All of them are polyphenols: flavonoids, flavonol or flavones. The best synthetic products in this ranking were Lopinavir and Nelfinavir (rank 7 and 10).
Table 4: Rank of compounds regard to Lupinski's rules violation. |
|||
| Name | Type | Rank | Violation |
| Nictoflorin | P | 1 | 3 |
| Quercitrin | P | 2 | 2 |
| Luteolin-7-Glucoside | P | 3 | 2 |
| Gossypetin-3'-O-Glucoside | P | 4 | 2 |
| Gossypetin-7-O-Glucoside | P | 5 | 2 |
| Gossypetin-8-Glucoside | P | 6 | 2 |
| Lopinavir | D | 7 | 1 |
| Astragalin | P | 8 | 2 |
| Gossypetin-3-Glucoside | P | 9 | 2 |
| Nelfinavir | D | 10 | 1 |
| Piceid | P | 11 | 1 |
| Remdesivir | D | 12 | 2 |
| Kaempferol | P | 13 | 0 |
| Quercetin | P | 14 | 0 |
| Natural Ligand |
|
15 | 2 |
| Aloenin | P | 16 | 0 |
| Amiodarone | D | 17 | 2 |
| Arbidol | D | 18 | 1 |
| Curcumin | P | 19 | 0 |
| Digitoxigenin | P | 20 | 0 |
| Ribavirin | D | 21 | 0 |
| Crocin | P | 22 | 3 |
| Hydroxychloroquine | D | 23 | 0 |
| Cannabidiol | P | 24 | 1 |
| Chloroquine | D | 25 | 0 |
| Beta-Eudesmol | P | 26 | 0 |
G3'G and derivatives docking
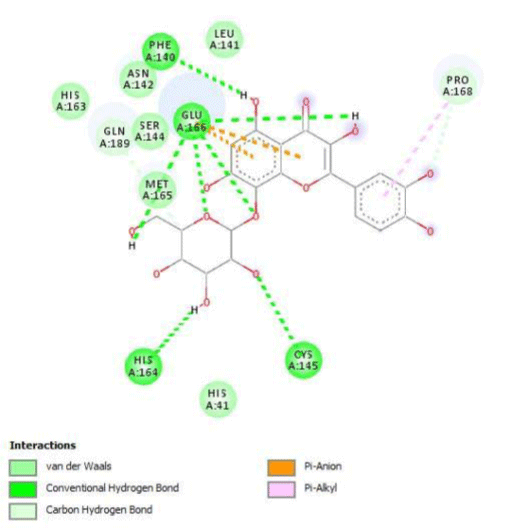
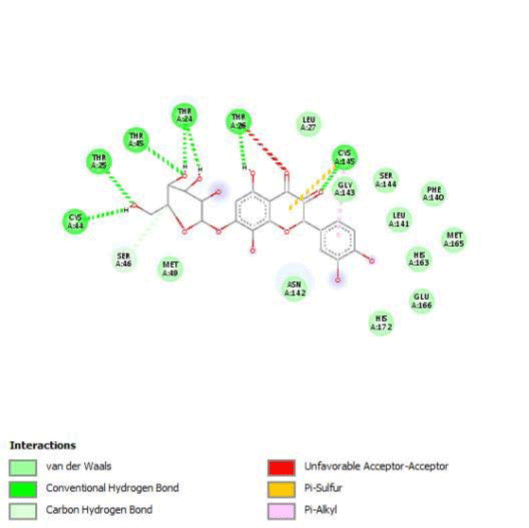
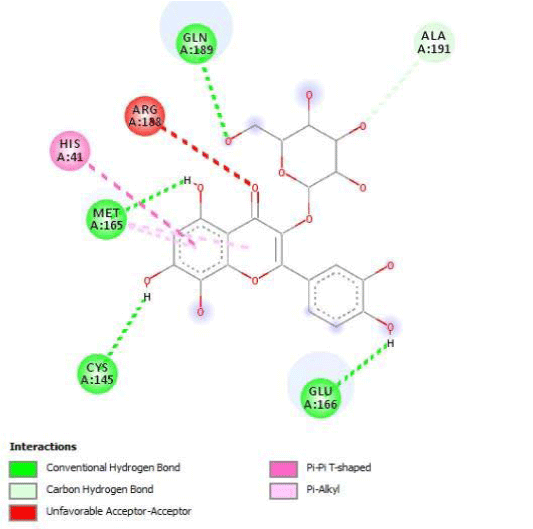
G3'G and other gossypetin derivates link at least one of the essential AA of binding site domains I and II of the catalytic center: Cys145 or His41. All others AA of the active site are brought into play according to the ligand but not all together. G3G and G8G are the only ligands that bind critical conserved His41 as well as Cys145. Glu166 is a recurrent linker through all gossypetin and derivatives.
This residue belongs to the sub-sites S1 to S5
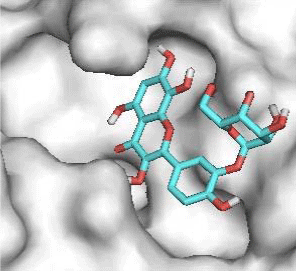
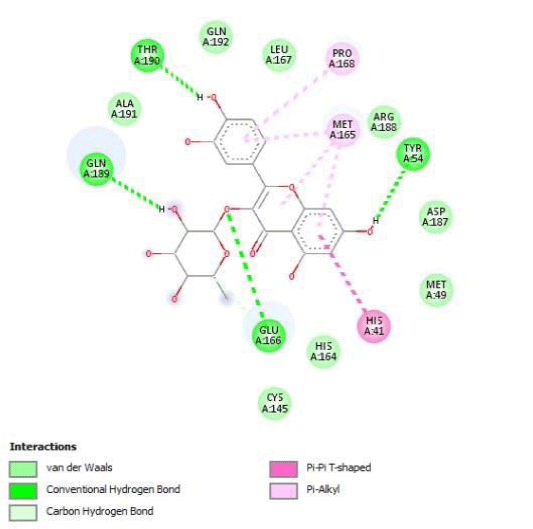
Gossypetin-8-Glucoside (G8G) and Gossypetin-7-O-Glucoside (G7G) presents the most bonds to the protein. They established thirteen bonds of several types. All the residues involved in the links with G8G are known residues of the active site. G7G links probably new and non-critical residues: Thr24, Thr26, Thr45, Cys44, Ser46 and His163.
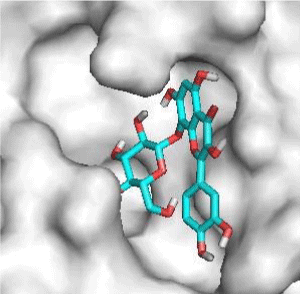
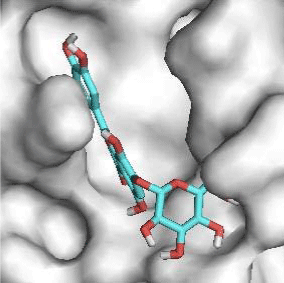
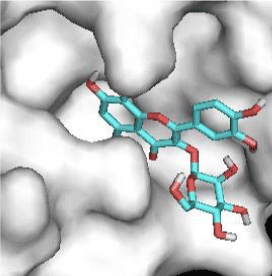
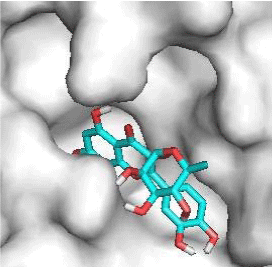
Quercitrin, G3'G and Gossypetin-3-Glucoside (G3G) establish respectively 9, 8 and 7 bonds with known residues of active site. We found that Quercitrin links 3 times with Met165. Figure 2 summarized all of those data.
We develop a computational method of prediction of best candidate inhibitors of SARS-CoV-2 3-Chymotrypsin-Like Protease (3CLpro) that plays an essential role in replication of the virus, and represents an effectiveness and nontoxic drug target. This computer-based analysis demonstrates for the first time, a potential benefit of G3'G and derivatives to fight COVID-19.
Covid-19 is a viral infection which fastly spread worldwide, and whose treatment remains an unresolved challenge so far. Among many drugs evaluated, few have proven conclusive clinical efficacy. Furthermore, the spread of the disease mandates that ideal medications against Covid-19 be cheap, and available worldwide. Therefore, there is a rationale to evaluate whether treatments of natural origin from aromatic and medicinal plants have the ability to prevent and/or treat COVID-19. Also, computation-based studies, which have proven efficacy in the screening of new active medications, fulfill the needs for fast analysis of any potential benefit.
Our demonstration of the benefit of Gossypetin derivatives follows a comprehensive, step by step process. First, we targeted 3CLpro, a protein-enzyme which plays a critical role in virus replication. Then, along with putative medicinal plants derived products, we included in our analysis molecules proposed in the Guidelines-version 6 for treatment of COVID-19. We also included compounds that have already been computationally tested. We experiment molecular docking methods which are widely used for this purpose. We used 3 methods: Autodock 4.2.6, AutodockVina version 1.1.2, and Smina version Oct 15 2019. Each method uses a different and inaccurate approximation for a given problem. Either program can provide a better result. For example, AutoDock is better in discriminating ligands and decoys in more hydrophobic, poorly polar and poorly charged pockets, while Vina tend to give better results for polar and charged binding Pockets [33]. So we compiled those results using a consensus scoring approach.
The computational study is based on a number of repetitions of tests and the compilation of 3 prediction methods. In order to propose the most representative score docking of molecules, we perform fifty runs per ligand with Autodock 4.2.6 and Smina method and the maximum of twenty runs performed for AutodockVina. The ranking of candidates on the basis of free binding energy with respect to violations of Lipinski's rules and other physicochemical binding properties is common but probably not sufficient to offer ready-to-use molecules.
But a deep inside comparative study of Traditional Chinese Medecine versus conventional medicine tend to prove the efficacy of using natural compounds by traditional habits [4,34]
Quercitrin and some of its derivatives such as Nictoflorin, Luteolin-7-glucoside or Astragalin, and all Gossypetin derivatives have the best free binding energy. According to our study, they are good therapeutic candidates. However, Nictoflorin presents 3 violations of Lupinski's rules whereas all the others present only 2. This reduces Nictoflorin claims of oral absorption. So therefore, Gossypetins remain the most likely natural compounds candidate for treatment of COVID-19. Their inhibition constant, their dissolving energy and the quantity of predicted H-bonds are among the best of our dataset.
Potential of G3'G and derivatives as a Covid-19 treatment, is reinforced by the visualization of molecular interactions between the active site of 3CLpro/all Gossypetin derivatives. Those interactions, rendered possible by our successful 3-D modeling of Gossypetin-3'-O-glucoside (G3'G), shows that all these molecules interact with AA of the catalytic center. Glu166, which belongs to the S1 to S5 sub-sites, is systematically bounded. And G3G and G8G bind the critical conserved His41 and Cys145 residues of the catalytic center.
For all of those reasons we propose G8G, G3G, G3'G and finally G7G as good tropical natural compounds candidate that should be further investigated to prevent or treat COVID19. We were able to show that molecules derived from medicinal plants have better physico-chemical interaction properties and potential for inhibition of 3CLpro, as compared to some candidate drugs proposed for the treatment of COVID19.
Our computation analysis matches with previous analyses. We were able to verify that according to their ΔG, Digitoxigenin>Beta-eudesmol, as suggested by Aanouz et al. Our method does not allow us to compare Crocin with them. Similarly, in accordance with the experience of Khaerunnisa et al., Nelfinavir/Lopinavir>Kaempferol/Quercetin. However our classification methods propose a lower consensus-score of ΔG for Luteolin-7-Glucoside. Several reasons can explain this difference: 1. we worked with protonated protein and ligands, 2. The coordinates of the grid are generally not communicated by others, and probably significantly different from one experiment to another, 3. Our method for minimizing the ligand are different and the choice of conformer used for experiments may vary from one experimenter to another. Martinique is a tropical caribbean island. Biodiversity is a pride and a wealth. The population still practices traditional medication with the use of local medicinal plants.
Only four flavonoid glycosides have been found in nature derivatives from gossypetin: G8G or gossypin (H. vitifolius and H. sabdariffa); G7G or gossypitrin/gossypetrin (H. sabdariffa, T. elatum and T. tiliaceum); G3G or gossytrin (H. sabdariffa and T. tiliaceum) and G3'G (A. manihot and T. elatum). For the first time G3'G was extracted and characterized from petals of flowers of Talipariti elatum Sw. of Martinica [17]. The high solubility of G3'G in the aqueous phase makes it, like many other molecules derived from medicinal plants, a good candidate for treatments. Its antifungal and antioxidant properties are already known. His anti-SARS-COV-2 properties have never been explored.
The computational study that we have carried out confirms the interest of polyphenols in the treatment of SARS-COV2 by inhibition of 3CLpro. In particular, the Gossypetin derivatives present in tropical Hibiscus varieties are for the 1st time classified according to a predictive in sillico method, on the same level as Quercitrin or luteolin-7-Glucoside proposed by Indian research teams, and present a priori a better inhibitory potential than Lopinavir / Nelfinavir treatments against 3CLpro. The computational study that we have carried out confirms the interest of flavonoids in the treatment of SARS-COV-2 by inhibition of 3CLpro. It encourages research on local medicinal plants for the treatment of COVID-19. In particular, the secondary metabolites of Gossypetin extracted from tropical varieties of Hibiscus appear promising. Like all computational studies, our study should be supplemented with in vivo experiments to refine the therapeutic proposals. It could be the support for proposing local therapeutic or preventive trials and epidemiologic studies.
To go further in proposing new treatments, the molecules derived from medicinal plants seem interesting. We could broaden our screening of local medicinal plants compounds candidates. We could target another SARS-COV2 element such as other enzymes or specific structural proteins. Molecular dynamic study could also be the next step to confirm results or evaluate combination of therapeutics. In vivo experiments remain essential to confirm therapeutic efficacy.
Jocelyn INAMO, giving us valuable reviewing on discussion and Max MONAN advises for information about Gossypetins molecules and molecular chemistry.
- Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009 Mar;7(3):226-36. doi: 10.1038/nrmicro2090. Epub 2009 Feb 9. PMID: 19198616; PMCID: PMC2750777.
- Khan SA, Zia K, Ashraf S, Uddin R, Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J Biomol Struct Dyn. 2020 Apr 13:1-10. doi: 10.1080/07391102.2020.1751298. Epub ahead of print. PMID: 32238094.
- Lin Q, Zhao S, Gao D, Lou Y, Yang S, Musa SS, Wang MH, Cai Y, Wang W, Yang L, He D. A conceptual model for the coronavirus disease 2019 (COVID-19) outbreak in Wuhan, China with individual reaction and governmental action. Int J Infect Dis. 2020 Apr;93:211-216. doi: 10.1016/j.ijid.2020.02.058. Epub 2020 Mar 4. PMID: 32145465; PMCID: PMC7102659.
- Chen Z, Nakamura T. Statistical evidence for the usefulness of Chinese medicine in the treatment of SARS. Phytother Res. 2004 Jul;18(7):592-4. doi: 10.1002/ptr.1485. PMID: 15305324.
- Tahir Ul Qamar M, Alqahtani SM, Alamri MA, Chen LL. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. 2020 Aug;10(4):313-319. doi: 10.1016/j.jpha.2020.03.009. Epub 2020 Mar 26. PMID: 32296570; PMCID: PMC7156227.
- Anand K, Yang H, Bartlam M, Rao Z, Hilgenfeld R. Coronavirus main proteinase: target for antiviral drug therapy. Coronaviruses with Special Emphasis on First Insights Concerning SARS. 2005:173–99. doi: 10.1007/3-7643-7339-3_9. PMCID: PMC7123552.
- Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013 Dec;4(12):951-61. doi: 10.1007/s13238-013-3096-8. Epub 2013 Dec 8. PMID: 24318862; PMCID: PMC4875403.
- Naksuk N, Lazar S, Peeraphatdit TB. Cardiac safety of off-label COVID-19 drug therapy: a review and proposed monitoring protocol. Eur Heart J Acute Cardiovasc Care. 2020 Apr;9(3):215-221. doi: 10.1177/2048872620922784. Epub 2020 May 6. PMID: 32372695; PMCID: PMC7235441.
- Ruamviboonsuk P, Lai TYY, Chang A, Lai CC, Mieler WF, Lam DSC; for Asia-Pacific Vitreo-Retina Society. Chloroquine and Hydroxychloroquine Retinal Toxicity Consideration in the Treatment of COVID-19. Asia Pac J Ophthalmol (Phila). 2020 Mar-Apr;9(2):85-87. doi: 10.1097/APO.0000000000000289. PMID: 32349115; PMCID: PMC7227199.
- S Khaerunnisa, H Kurniawan, R Awaluddin, S Suhartati, S Soetjipto. Potential Inhibitor of COVID-19 Main Protease (Mpro) From Several Medicinal Plant Compounds by Molecular Docking Study. Medicine & Pharmacology, preprint, mars. 2020. doi: 10.20944/preprints202003.0226.v1.
- AK Srivastava, A Kumar, N Misra. On the Inhibition of COVID-19 Protease by Indian Herbal Plants: An In Silico Investigation. arXiv. 2020. https://tinyurl.com/y5u8umsd
- Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov Ther. 2020;14(1):58-60. doi: 10.5582/ddt.2020.01012. PMID: 32147628.
- Wang G, Zhu W. Molecular docking for drug discovery and development: a widely used approach but far from perfect. Future Med Chem. 2016 Sep;8(14):1707-10. doi: 10.4155/fmc-2016-0143. Epub 2016 Aug 31. PMID: 27578269.
- Aanouz I, Belhassan A, El-Khatabi K, Lakhlifi T, El-Ldrissi M, Bouachrine M. Moroccan Medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. J Biomol Struct Dyn. 2020 May 6:1-9. doi: 10.1080/07391102.2020.1758790. Epub ahead of print. PMID: 32306860; PMCID: PMC7212546.
- Joshi T, Joshi T, Sharma P, Mathpal S, Pundir H, Bhatt V, Chandra S. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. Eur Rev Med Pharmacol Sci. 2020 Apr;24(8):4529-4536. doi: 10.26355/eurrev_202004_21036. PMID: 32373991.
- GA Gyebi, OB Ogunro, AP Adegunloye, OM Ogunyemi, SO Afolabi. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CL pro ): An in silico screening of alkaloids and terpenoids from African medicinal plants. J Biomol Struct Dyn. 2020;1‑13. doi: 10.1080/07391102.2020.1764868.
- Frantz François-Haugrin, Max Monan, Emmanuel Nossin, Juliette Smith-Ravin, Odile Marcelin. Antioxidant activity of an isomer of gossypitrin (gossypetin-3'-O-glucoside) isolated in the petals of Talipariti elatum Sw., and determination of total phenolic content of the total flower. J Pharmacogn Phytochem 2016;5(5):200-208. https://tinyurl.com/yybspabz
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. doi: 10.1093/nar/28.1.235. PMID: 10592235; PMCID: PMC102472.
- Xu Z, Peng C, Shi Y, Zhu Z, Mu K, Wang X, Zhu W. Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. bioRxiv; 2020. doi: 10.1101/2020.01.27.921627.
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y, Yu J, Wang L, Yang K, Liu F, Jiang R, Yang X, You T, Liu X, Yang X, Bai F, Liu H, Liu X, Guddat LW, Xu W, Xiao G, Qin C, Shi Z, Jiang H, Rao Z, Yang H. The crytal structure of 2019-nCoV main protease in complex with an inhibitor N3. Nature. 2020;582:289-293. https://tinyurl.com/yyw4p86s
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH. PubChem Substance and Compound databases. Nucleic Acids Res. 2016 Jan 4;44(D1):D1202-13. doi: 10.1093/nar/gkv951. Epub 2015 Sep 22. PMID: 26400175; PMCID: PMC4702940.
- Lowe DM, Corbett PT, Murray-Rust P, Glen RC. Chemical name to structure: OPSIN, an open source solution. Journal of Chemical Information and Modeling. 2011 Mar;51(3):739-753. DOI: 10.1021/ci100384d.
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform. 2011 Oct 7;3:33. doi: 10.1186/1758-2946-3-33. PMID: 21982300; PMCID: PMC3198950.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001 Mar 1;46(1-3):3-26. doi: 10.1016/s0169-409x(00)00129-0. PMID: 11259830.
- Huang B. MetaPocket: a meta approach to improve protein ligand binding site prediction. OMICS. 2009 Aug;13(4):325-30. doi: 10.1089/omi.2009.0045. PMID: 19645590.
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W665-7. doi: 10.1093/nar/gkh381. PMID: 15215472; PMCID: PMC441519.
- MHM Olsson, CR Sondergaard, M Rostkowski, JH Jensen. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical p Ka Predictions. J Chem Theory Comput. 2011;525‑537. doi: 10.1021/ct100578z.
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009 Dec;30(16):2785-91. doi: 10.1002/jcc.21256. PMID: 19399780; PMCID: PMC2760638.
- Xu W, Lucke AJ, Fairlie DP. Comparing sixteen scoring functions for predicting biological activities of ligands for protein targets. J Mol Graph Model. 2015 Apr;57:76-88. doi: 10.1016/j.jmgm.2015.01.009. Epub 2015 Feb 1. PMID: 25682361.
- Quiroga R, Villarreal MA. Vinardo: A Scoring Function Based on Autodock Vina Improves Scoring, Docking, and Virtual Screening. PLoS One. 2016 May 12;11(5):e0155183. doi: 10.1371/journal.pone.0155183. PMID: 27171006; PMCID: PMC4865195.
- Koes DR, Baumgartner MP, Camacho CJ. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J Chem Inf Model. 2013 Aug 26;53(8):1893-904. doi: 10.1021/ci300604z. Epub 2013 Feb 12. PMID: 23379370; PMCID: PMC3726561.
- Wang R, Wang S. How does consensus scoring work for virtual library screening? An idealized computer experiment. J Chem Inf Comput Sci. 2001 Sep-Oct;41(5):1422-6. doi: 10.1021/ci010025x. PMID: 11604043.
- TF Vieira, SF Sousa. Comparing AutoDock and Vina in Ligand/Decoy Discrimination for Virtual Screening. Appl Sci.2019;4538. doi: 10.3390/app9214538.
- Y Yang, MS Islam, J Wang, Y Li, X Chen. Traditional Chinese Medicine in the Treatment of Patients Infected with 2019-New Coronavirus (SARS-CoV-2): A Review and Perspective. Int J Biol Sci. 2020;1708‑1717. doi: 10.7150/ijbs.45538.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.