> Biology. 2020 August 12;1(4):070-087. doi: 10.37871/jels1123.
-
Subject area(s):
- Infectious Diseases
- Virology
- Antivirology
- Antiretrovirology
Covid-19 Pandemic-Insights and Challenges
Lakshmi S*, Shehna S, Vimal S, Midhu GV, Shiny DV, Sreelekshmi S, Reshmi R and Abi SA
#All authors have equally contributed in the framing of manuscript
- Middle East respiratory syndrome
- Pathophysiology
- Covid-19
- Betacoronavirus
- Severe acute respiratory syndrome
- Acute respiratory distress syndrome
The 2019 Novel Coronavirus (2019-nCoV) outbreak affected a large number of deaths with millions of confirmed cases worldwide. Coronavirus Disease (COVID-19) is associated with respiratory illness that lead to severe pneumonia and Acute Respiratory Distress Syndrome (ARDS). Although related to the Severe Acute Respiratory Syndrome (SARS) and the Middle East Respiratory Syndrome (MERS), Covid-19 shows some unique pathogenetic, epidemiological and clinical features. On the basis of the phylogenetic relationship as well as genomic structures, the Covid-19 belongs to genera Betacoronavirus. Human Betacoronaviruses (SARS-CoV-2, SARS-CoV, and MERS-CoV) have shared similarities, yet differences also in their genomic and phenotypic level that influence the pathogenesis. To gain knowledge regarding the pathophysiology and virulence of the Covid-19 virus, it is absolutely necessary to understand its genetic makeup, transmission, virulence factors, risk factors, diagnosis, clinical presentations, outcome predictions, management of risk factors and ways to control the disease thus providing an insight to the current or future treatment and management protocols. To provide a review of the differences in pathogenesis, epidemiology and clinical features of Covid-19, its transmission and replication dynamics, genome organization, current clinical trials and vaccine development strategies, Immunoinformatics, diagnostics and ways to control the pandemic, inorder to raise an increasing awareness, both to the public and for scientific perspectives.
Coronaviruses belong to large family of enveloped positive sense RNA viruses which is surrounded by crown shaped, club like spike projections on the outer surface which are responsible for respiratory illness in humans and animals. The 2019 Novel Coronavirus (2019-nCoV; Family Coronaviridae), also termed as the Wuhan coronavirus, has created a global emergency which had not even seen in the 2003 Severe Acute Respiratory Syndrome (SARS) outbreak. It rapidly spread spanning the entire area, causing an epidemic in China, followed by an increasing number of cases in other countries across the globe resulting in a pandemic situation. In February 2020, the World Health Organization termed the pandemic as "COVID-19", which stands for coronavirus disease 2019. The virus was first identified in Wuhan city of Hubei province of china that leads to a contagious, respiratory infection, with an incubation of about 10 days. Most of the studies indicated the possibility of transmission which could also be asymptomatic for several days. Similar to other coronaviruses, 2019-nCoV reportedly possesses a surface glycoprotein. In more established coronavirus pathogenesis, this protein has been shown to bind host cellular receptors and to mediate membrane fusion. More importantly, this has been described as a potential vaccine target in both Sars Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [1].
Genome sequencing and phylogenic analysis indicated that the Coronavirus that causes COVID-19 is a betacoronavirus in the same subgenus as the Severe Acute Respiratory Syndrome (SARS) virus but in a different clade. The receptor-binding gene region of Covid-19 is very similar to that of the SARS coronavirus, and the virus has been shown to use the same receptor, the Angiotensin-Converting Enzyme 2 (ACE2), for cell entry [2-4]. The Coronavirus Study Group of the International Committee on Taxonomy of Viruses has proposed that this virus be designated Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [5]. The MERS virus, yet another betacoronavirus, appears more distantly related [6,7]. The closest RNA sequence similarity is to two bat coronaviruses, and it appears likely that bats are the primary source; whether Covid-19 virus is transmitted directly from bats or through the intermediate system is still unknown [8].
In a phylogenetic analysis of various strains of SARS-CoV-2 from China, two different types of SARS-CoV-2 were identified, designated type L (accounting for 70 percent of the strains) and type S (accounting for 30 percent) [9]. The geographical spread is about more than three million confirmed cases reported while the cumulative incidence varies by circumstances based on a number of factors, including population density and demographics, extent of testing and reporting, and timing of mitigation strategies. In the United States, outbreaks in long-term care facilities and homeless shelters have emphasized the risk of exposure and infection in congregate settings. The transmission outbreak progressed, person-to-person spread became the main mode of transmission.
Period of infectivity had become debatable as SARS-CoV-2 can be transmitted prior to the development of symptoms and throughout the course of illness. The risk of transmission from an individual with SARS-CoV-2 infection varies by the type and duration of exposure, use of preventive measures, and likely considerable factors. Most secondary infections have been described among by the person to person contact settings in health care and other areas [9,10]. Clusters of cases have also been reported in family, work, or social gatherings where close and personal contact occur. The indirect contact like passing someone with infection on the street, handling items that were previously handled by someone with infection is not well confirmed but low. The contaminated surfaces may be another source of infection if susceptible individuals touch these surfaces and then transfer infectious virus to mucous membranes through the mouth, eyes, or nose. It may be more likely to be a potential source of infection where there is heavy viral load.
There has been a great deal of Covid-19 research, involves accelerated processes and publishing shortcuts to meet the global demand. Here we aim to review the past, present and future of the subgenus Covid-19 of coronaviridae family for a better understanding so as to evolve new breakthrough for its containment.
Evolving status of Covid-19
Coronaviruses (CoV) are generally large family of viruses that belongs to the Coronaviridae family of order Nidovirales, which mainly causes infections in the respiratory and gastrointestinal tract. Based on phylogenetic clustering, the subfamily Orthocoronavirinae are divided into four genera as; alpha, beta, gamma and delta-coronavirus. The alpha and beta-coronaviruses were reported to infect mammals, whereas gamma and delta-coronaviruses infect birds [11]. CoV are positive-sense single stranded RNA viruses having a spike glycoprotein on the envelope, and appeared like a crown when viewed under an electron microscope.
Major pandemic, epidemic diseases in past 20 years includes: Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-1) in 2003, H1N1 influenza in 2009 (first influenza pandemic of the 21st century), the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in 2012, the Ebola epidemic in West Africa in 2014, and the Zika virus in 2015.
Severe Acute Respiratory Syndrome (SARS) first emerged in mid-November 2002 in the Guangdong Province in the southern part of China [12]. SARS was caused by a virus in the Coronavirus (CoV) family [13,14], which is a bat-based animal virus [15-18]. The initial cases were animal handlers in Guangzhou city, which was then spread rapidly and globally after a period of three months through person-to-person in Hong Kong and then to other countries including United States of America, Vietnam, Singapore and Canada. The infected patients showed symptoms of pneumonia with a diffused alveolar injury and ARDS [19], eventually leading to pulmonary failure and fatality. A total of 8422 probable cases of SARS were reported in 32 countries with 916 deaths (11% case-fatality rate), during the period from November 1, 2002 to August 7, 2003 [20].
Ten years after the SARS outbreak, another human coronavirus known as Middle East Respiratory Syndrome Coronavirus (MERS-CoV) emerged in June of 2012 in the Middle Eastern Countries. It was first identified in Saudi Arabia. MERS-CoV marked the second zoonotic coronavirus infection affecting human, probably originating from bats [21]. But some data provided the major reservoir of MERS as dromedary camels [22,23], however the exact role of dromedaries and the exact route of virus transmission were unknown. Soon after, the World Health Organization issued a global alert. The common symptoms of MERS included fever, cough and shortness of breath. Pneumonia was also a minor symptom. Gastrointestinal symptoms, including diarrhea, have also been reported. MERS-CoV infection showed initiates from a mild upper respiratory injury to severe acute pneumonia, rapidly progressing to Acute Lung Injury (ALI), ARDS, and multi-organ failure resulting in death [24]. Since September 2012, twenty seven countries have reported MERS-CoV cases to WHO with a total of 2494 laboratory-confirmed cases, including 858 associated deaths (case-fatality rate: 34.4%) [25]. MERS-CoV was considered as a notifiable disease under the International Health Regulations (2005).
Covid-19 is a new coronavirus that emerged in late December of 2019 in Wuhan City, Hubei Province of China, which have not seen in humans before. The National Health Commission of China reported that outbreak is associated with exposures in one seafood market in Wuhan city. International Committee on Taxonomy of Viruses (ICTV) termed the outbreak disease as SARS-CoV-2, due to similarity of its symptoms to those induced by SARS. Covid-19 is neither SARS nor influenza. It is a new virus with its own unique characteristics which is highly contagious, spreading quickly and is considered capable of causing enormous health, economic and societal impacts. Covid-19 is a zoonotic virus, which implies that they first developed in animals before developing in humans. Studies are still in progress to understand the zoonotic origin of this outbreak. As the animal origin of the virus is unknown at present, the possibility of reappearance of the disease into previously infected areas are constantly considered.
Infected people show symptoms of fever, dry cough, fatigue, sputum production, shortness of breath/dyspnoea, myalgia or arthralgia [26], sore throat and headache. 5% of patients were reported to have nausea or vomiting also [27]. According to Centers for Disease Control and Prevention (CDC), the symptoms appeared 2-14 days after exposure to the virus, and the full list of symptoms are still being investigated. Most serious complication of Covid-19 is a type of pneumonia that's been called Novel Covid-19 Infected Pneumonia (NCIP). Other complications of the infection included ARDS, RNAemia, acute cardiac injury, other associated secondary infections [26], septic shock, respiratory failure, arrhythmia, liver dysfunction, multi-organ failure eventually resulting in death.
Covid-19 virus has been detected in respiratory, fecal and blood specimens [27]. Virus could spread through respiratory droplets expelled during cough or sneeze and are then transmitted during close unprotected contact between an infector and infected. Viable virus was identified from fecal shedding of some patients, and in some cases live virus had been cultured from infected person's stools. But the fecal-oral route of transmission is yet to be determined and understood [26]. The air-borne spread of Covid-19 has not been reported to date. Different stages of Virus Transmission can be categorized into four as; (i) Stage 1: First appearance of the disease, (ii) Stage 2: Local Transmission, (iii) Stage 3: Community Transmission, and (iv) Stage 4: Widespread Outbreak.
Covid-19 virus doesn't replicate by themselves, rather use host genome machinery to replicate. Its genome has an overall identity of 96% with bat SARS-like coronavirus, 86%-92% to a pangolin SARS-like coronavirus [27], 82% with that of human SARS-CoV [28] and shares greater than 99.9% sequence identity, indicating a recent host shift into humans [29]. The epidemiology of Covid-19 is rapidly evolving when people mix together; which happens in workplace, households, and on the journeys people make. As of 07 May 2020, there have been 3,595,662 confirmed cases of Covid-19 reported to WHO in 215 countries, areas or territories, including 247,652 deaths [30-40]. Table 1 illustrates events of Covid-19 pandemic throughout the world.
| Table 1: Events of outbreaks of Covid-19 pandemic across the globe. | ||
| Date | Key Events | References |
| December 31, 2020 | An outbreak of pneumonia of unknown etiology was detected in Wuhan [China] and reported to WHO Country Office in China | [31] |
| January 03, 2020 | Information on the epidemic was notified to WHO | [27] |
| January 04, 2020 | China reported a cluster of pneumonia cases to WHO with no deaths in Wuhan | [31] |
| January 07, 2020 | The etiological agent of the atypical pneumonia was isolated from sea food market in Wuhan city by the Chinese research authorities and named as novel coronavirus [2019-nCoV] | [32] |
| January 10, 2020 | The whole genome sequences of the COVID-19 viruses were shared with WHO and the international community | [27] |
| January 12, 2020 | The genetic sequence of the novel coronavirus identified was shared by China to other countries to develop specific diagnostics kits | [31] |
| January 13, 2020 | First case of novel coronavirus outside of China was confirmed. Ministry of Public Health, Thailand reported their first imported case of lab-confirmed 2019 n-CoV from Wuhan, China | [31,32] |
| January 15, 2020 | Ministry of Health, Labour and Welfare, Japan [MHLW] reported their first imported case of lab-confirmed 2019 n-CoV from Wuhan, China | [32] |
| January 20, 2020 | COVID-19 was included in the statutory report of Class B infectious diseases and border health quarantine infectious diseases National IHR Focal point from the Korea reported their first imported case of lab-confirmed 2019 n-CoV United States confirmed first case of 2019-nCoV |
[32]
[32] [33] |
| January 21, 2020 | The Chinese government shared information regarding viral sequences, the PCR primers and probes used in the rRT-PCR test kit with WHO and the international community. | [31,27] |
| January 24, 2020 | Vietnam has reported first case of 2019-nCoV with no travel history from China, while his family member was the China traveler. Hence marks the first incidence of human to human transmission of corona virus The government of Singapore confirmed first case of 2019-nCoV |
[34] |
| January 25, 2020 | The government of Australia, federal democratic republic of Nepal and French republic confirmed their first cases of 2019-nCoV | [35] |
| January 26-31, 2020 | Other countries reported the cases of 2019-nCoV; Jan 26 [Malaysia], Jan 27 [Canada], Jan 28 [Cambodia, Germany, Sri Lanka], Jan 29 [United Arab Emirates], Jan 30 [Philippines, India, Finland], and Jan 31 [Italy] | [36-41] |
| January 30, 2020 | WHO declared the outbreak a Public Health Emergence of International Concern [PHEIC] under the International Health Regulations 2005 | [31] |
| February 01, 2020 | Russian Federation, Spain, Sweden, and United Kingdom reported the cases of 2019-nCoV | [42] |
| February 05, 2020 | Belgium reported the case of 2019-nCoV | [43] |
| February 11, 2020 | WHO announced a name for the disease caused by new virus as a "COVID-19" which is the acronym of "Coronavirus Disease 2019" | [31] |
| February 15, 2020 | Egypt reported the case of 2019-nCoV | [44] |
| February 19, 2020 | Islamic Republic of Iran reports the first confirmed case of COVID-19 | [45] |
| February 22, 2020 | Lebanon and Israel reported the cases of COVID-19 | [46] |
| March 06, 2020 | Bhutan, Cameroon, Serbia, and South Africa reported the cases of COVID-19 | [47] |
| March 07, 2020 | 100 countries are reporting 100, 000 cases of COVID-19. | [31] |
| March 11, 2020 | WHO declared COVID-19 as pandemic | [31] |
| March 13, 2020 | Europe becomes the epicenter of the pandemic, with more reported cases and deaths, apart from China | [31] |
| March 16, 2020 | Phase I clinical trial to investigate mRNA-1273, a vaccine developed by Moderna against the COVID-19 has begun in Kaiser Permanente Washington Health Research Institute [KPWHRI], Seattle, US. Jennifer Haller [44] is the first person [participant] in world to get coronavirus vaccine trial | [48,49] |
| March 18, 2020 | WHO and partners launch the SOLIDARITY clinical trial for COVID-19 treatment | [31] |
| March 25, 2020 | Off-label use of medicines for the treatment of COVID-19 | [31] |
| March 28, 2020 | The U.S. Food and Drug Administration issued an Emergency Use Authorization [EUA] to allow hydroxychloroquine sulfate and chloroquine phosphate products in treating patients with COVID-19 | [50] |
| April 13, 2020 | Under WHO's coordination, expert group forms for the development of vaccines against COVID-19 | [31] |
| April 21, 2020 | The Food and Drug Administration [FDA] has approved the use of the first COVID-19 at-home test kit, developed by LabCorp | [51] |
Diagnosis and monitoring of infection
Suspicion and criteria: The incidence of Covid-19 should be firstly considered in candidates with onset fever and respiratory tract difficulties along with myalgias, diarrhea, and smell or taste aberrancies. The likelihood of Covid-19 is increased if the patient: A resided or travelled to a location where there is community transmission of severe acute respiratory syndrome coronavirus b had close contact with a confirmed or suspected case of Covid-19 or having direct contact with infectious secretions while not wearing Personal Protective Equipment (PPE), [41-46]. The diagnosis could be definitively made with microbiologic testing, testing criteria is as suggested by the World Health Organization (WHO), because of the limited availability of testing, the diagnosis of Covid-19 is made presumptively based on clinical presentation on the basis of exposure risk.
Techniques
Viral testing by RT-PCR: The diagnosis of COVID-19 is made by detection of SARS-CoV-2 RNA by Reverse Transcription Polymerase Chain Reaction (RT-PCR) [47-52]. Various RT-PCR assays are used around the world; different assays amplify and detect different regions of the SARS-CoV-2 genome. Common gene targets include Nucleocapsid (N), Envelope (E), Spike (S), and RNA-Dependent RNA Polymerase (RdRp), as well as regions in the first open reading frame [53].
Specimen collection: Upper respiratory samples are the primary specimens for SARS-CoV-2 RT-PCR testing. Oropharyngeal, nasal mid-turbinate, or nasal swabs are collected from the symptomatic patients in the absence of nasopharyngeal swabs. Nasal mid-turbinate or nasal swabs could be self-possessed by the patients [54]. Expectorated sputum and bronchoalveolar lavage should be considered from patients with productive cough and who are intubated.
Interpretation: A positive test for SARS-CoV-2 generally confirms the diagnosis of Covid-19. As the documentation of false-negative tests from upper respiratory specimens exist, the suspicion for Covid-19 remains that the test should be repeated by testing lower respiratory tract specimens as per the WHO [55]. Infection control precautions for Covid-19 should continue while repeat evaluations are being performed. The accuracy and predictive values of SARS-CoV-2 tests have not been systematically evaluated, and the sensitivity of testing likely depends on the precise RT-PCR assay, the type of specimen obtained, the quality of the specimen, and duration of illness at the time of testing.
The chance of a positive upper respiratory RT-PCR may be higher early during the duration of illness. Recent combinatorial study of RT-PCR and an Immunoglobulin (Ig)M serologic test to make the diagnosis of Covid-19 suggested that RT-PCR positivity rates were >90 percent on early days and later get decreased, so had to be interpreted with caution, due to the higher chances of false positivity [56].
Lower respiratory tract specimens could have higher viral loads that yielded positive tests than those obtained with the upper respiratory tract specimens [57,58]. In a study of 205 patients with Covid-19 who were sampled at various sites, the highest rates of positive viral RNA were demonstrated from bronchoalveolar lavage (95%) and sputum (72%) compared with oropharyngeal swab. Data supported the idea that viral RNA levels were higher and considerably detected in nasal part in comparison with oral specimens.
Serological studies for prior infection
The detection of antibodies to SARS-CoV-2 in the blood could be validated and could identify patients who have had Covid-19 or with current infections [59]. Detectable antibodies take time to develop. The study revealed that the Enzyme-Linked Immunosorbent Assay (ELISA) detected antibodies which could bind to the receptor-binding domain of the spike protein and was 12 days for IgM and 14 days for IgG, even though the rate of cross-reactivity with other coronaviruses would be a primary concern, also, IgM tests were prone to false-positive results [60].
Imaging: The predictable features regarding radiographs and the Computed Tomography (CT) of the chest of symptomatic reveals skewed peripheral ground-glass opacities without pleural effusions. Due to overlapping with the various other infections conditions such as adenovirus, imaging without confirmation by RT-PCR is of limited specificity [61,62].
Other tests: Several Tests that identify SARS-CoV-2 antigen are under development and as part of the safety reasons, specimens from a patient with documented Covid-19 was not allowed to be submitted for viral culture.
Immunoinformatics aided Identification of Covid-19
Current observations indicate that coronaviruses are particularly adapted to evade immune detection and dampen human immune responses. The prolonged incubation period is due to immune evasion properties, efficiently escaping host immune detection at the early stage of infection. As a member of the Betacoronavirus genus, immune evasion mechanism is potentially similar to those of SARS-CoV and MERS-CoV in which mechanisms rely on the repression of innate immune responses, especially type I interferon recognition and signaling [10]. The surface glycoprotein of 2019-nCoV have 76.3% identity and 87.3% similarity with the spike glycoprotein of SARS-CoV. The genomic data suggests SARS-CoV-2 and SARS-CoV has the corresponding human cell receptor, the Angiotensin-Converting Enzyme 2 (ACE2), while MERS-CoV uses Dipeptidyl Peptidase 4 (DPP4) for invasion [63]. The receptor affinity of ACE2 of SARS-CoV-2 is higher compared to SARS-CoV which will lead to severe lung involvement in Covid-19 than in SARS which requires further investigation.
MERS-CoV has progressed approach to manipulate innate immunity and prevent pathways leading to the production of IFN. This ability contributes to the higher rate of fatality in immunocompromised. The viral recognition by the TLR, recruit the adaptor molecules, either MyD88 (myeloid differentiation primary response 88) or Toll/Interleukin-1 Receptor- (TIR-) domain containing adapter-inducing interferon-β (TRIF) that further activate the pathway of MAPK and NF-κB responsible for the production of proinflammatory cytokines and IFNs [64-66]. The spike protein of MERS-CoV activate the expression of the negative regulators of the TLR signaling pathways which leads to the expression of IL-1R-Associated Kinase (IRAK-M) and Peroxisome Proliferator-Activated Receptor-γ (PPAR), the negative regulators of IRF7, the transcription factor that produces the expression of IFN-α and IFN-β [67]. The durable persistence of negative regulators reduces the clearance of MERS-CoV infections; thereby infection get established. The viral proteins including Membrane (M) or Nonstructural (NS) proteins (eg. NS4a, NS4b, NS15) are the key molecules in host immune modulation [68]. For adaptive immune evasion, antigen presentation via MHC class I and MHC class II will be down regulated when the macrophages or dendritic cells is infected with MERS-CoV, which would markedly diminish T cells activation [69,70].
Genomic organization of Covid 19
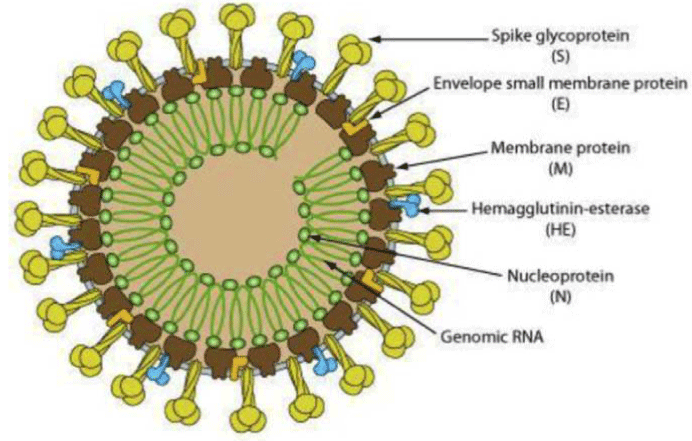
Covid-19 is a zoonotic RNA coronavirus, which is spherical or pleomorphic, non-segmented, single stranded, positive sense RNA [71] with 5' cap structure and 3' poly A tail virus. The total G+C content is 38%. The size of SARS-CoV 2 is 29.9 kb whereas SARS-CoV is 27.9 kb and MERS-CoV is 30.1 kb [72-75]. It has large genome of 29811 nucleotides [73] (cDNA) which includes 29.86% adenines, 18.39% cytosines, 19.63% guanines and 32.12% thymines [72]. Coronavirus belong to the order Nidovirales, in the family of Coronaviridae, subfamily of Coronavirinae. The subfamily includes 4 genera known as Alpacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus [73]. The alpha and beta genera infect mainly humans and mammals while the other 2 genera infect mainly birds. Novel coronavirus (SARS-Cov-2) belonging to beta genera is round shape with 60-140 nm diameter. A Virus caspid is comprised of matrix proteins and within the capsid, RNA associated with nucleoprotein is seen. Club-shaped glycoproteins are seen in virus envelope and a unique N-terminal fragment is seen within the spike proteins. Coronaviruses possess 32% to 43% variation in G+C contents [77]. In all coronaviruses, the genes that codes for structural proteins are in 5' to 3' order as S,E,M and N [77]. The Schematic representation of a Coronavirus is given as figure 1.
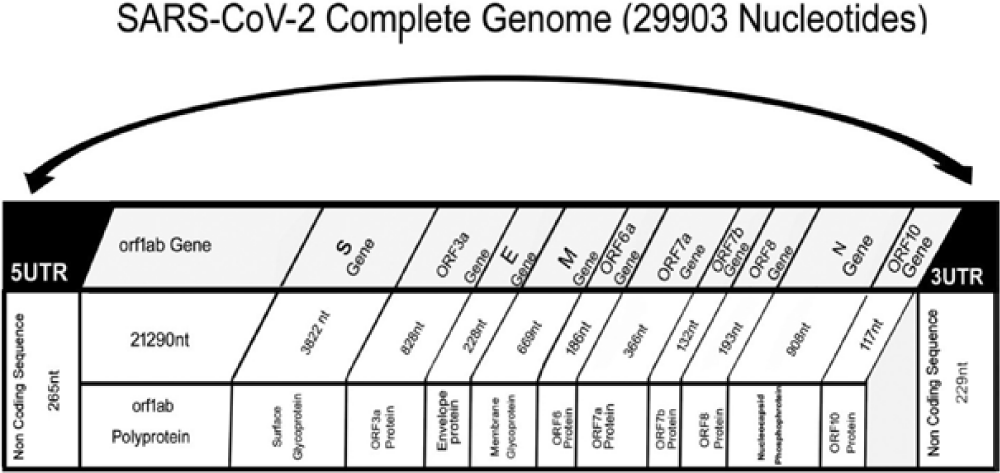
Coronaviruses contains at least six Open Reading Frames (ORFs) in their genome, there are few exceptions in case of Gammacoronavirus that lacks nsp1. Almost two thirds of the whole genome length is first ORFs (ORF1a/b) thatencodes 16 nsps. In Covid-19 there are 12 ORFs along with a set of 9 subgenomic mRNAs containing conserved leader sequence, nine transcription regulatory sequences and 2 terminal untranslated regions, but this virus don't have haemagglutinin-esterase gene which is found in lineage A beta-CoVs. The 16 nps are as follows: NSP3 (papanin like protease), NSP5 (main protease), NSP12 (RNA-dependent RNA polymerase, NSP13 (helicase) and other NSPs which are involved in transcription and replication of viruses [73]. Mutations have been observed in two NSPs such as NSP2 and NSP3 and spike proteins, which contribute to a major role in differentiation mechanism and infection of novel coronavirus, but at protein level, no aminoacid substitution was reported to occur in spike protein, NSP7, NSP13,envelop, accessory proteins p6 and 8b [74]. There are two Strains of Novel Coronavirus (SARS-CoV-2) which have been discovered such as L-type and S-type, L type is more aggressive and contagious which is derived from S-type [74]. The diversification degree of Covid 19 is smaller than mutation of H7N9 avian flu. The two polypeptides pp1a and pp1ab are produced by a frameshift in between ORF1a and ORF1b.Virally encoded Chymotrypsin-Like Protease (3CLpro) or main protease and 1 or 2 papain- like protease processes these polypeptides [75].
The four main structural proteins of the coronavirus contain spike, Membrane, envelope and nucleocapsid proteins represented as S,M,E and N respectively, encoded on one –third of the genome near the 3' terminus by ORF10 and ORF11. CoVs encode structural and accessory proteins such as HE protein, 3a/b protein and 4a/b protein which when matured play a major role in virus replication and genome maintenance [75]. Covid 19 virus genome contain 6 accessory proteins, encoded by ORF3a, ORF6, ORF7a, ORF7b and ORF8 genes [76]. M and E proteins helps in viral envelop formation while N protein mainly helps in assembly of the virus. The S structural protein bind specifically to the receptor of the host cell thereby helping viruses to invade susceptible cells in the host. Membrane Glycoprotein (M) constitute major portion of the structural protein, it spans the membrane bilayer three times, a long COO terminus (cytopasmic domain) is present inside the virion and short N2 terminal domain outside the virus [77]. Peplomers are Spike Proteins (S) as a type 1 membrane lycoproteins, S proteins are the main inducer of neutralizing antibodies. Intracellular viral particle formation is initiated by M glycoproteins without the involvement of S protein. Coronaviruses devoid of spike and are non-infectious can be grown in the presence of tunicamycin that contain M protein, but not contain S protein [77]. The Structure of the SARS-CoV-2 genome is given as figure 2.
Covid 19 shows certain genetic similarity with SARS-CoV (about 79%) and MERS-CoV (about 50%). The arrangement of N, E and M proteins are different among the Betacoronaviruses. The virus genomic RNA used to translate polyprotein 1a/1ab encodes Non Structural Proteins (NSPs) that leads to the formation of replication–transcription complex. It is formed inside the Double Membrane Vesicles (DMVs). Moreover, a nested set of Subgenomic RNAs (sgRNAs) are synthesized in a discontinuous transcription. These subgenomic mRNAs shares a common 5' leader and 3' terminal sequences. At transcription regulatory sequences there is transcription termination and subsequent acquisition of a leader RNA occurs which is located between ORFs and these Minus-Strand (sgRNAs) serve as template for subgenomic mRNAs production. The structural and accessory proteins are translated from sgRNAs [75].
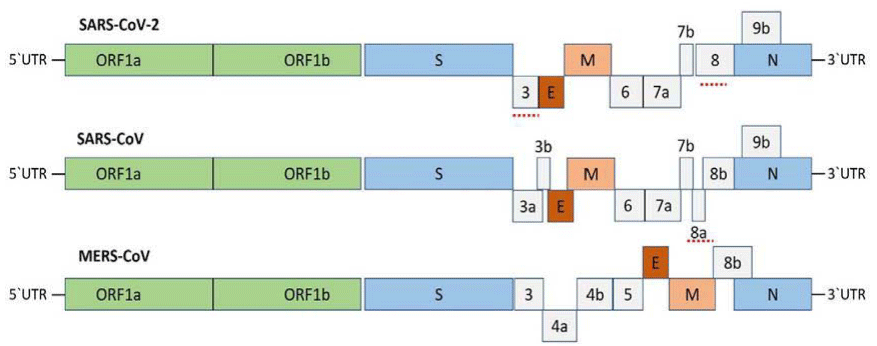
The nsp-coding region are more conserved and structural proteins are less conserved or more diverse which help them in adaptation inside the host species, which is proven from the genomic sequence alignment of Coronavirus which shows 58% identity on nsp-coding region and 43% on the structural protein-coding region among different coronaviruses. During the replication of RNA viruses, the mutation rates is more than that occurs in DNA viruses. The CoV genome is 30 kb in length (largest known RNA viruses) other RNA viruses genome having less than 10kb in length. The large genome of CoVs is nurtured with the help of special features of CoV RTC, which contains several RNA processing enzymes such as the 3'-5' exoribonuclease of nsp14. One of the unique feature of CoVs among all other RNA virus is this 3'-5' exoribonuclease activity, probably providing a proof reading function of the RTC. Sequence analysis of Covid-19 revealed that typical genomic structure of novel coronavirus belongs to the cluster of betacoronaviruses such as Bat-SL ZC45, SARS-CoV, Bat-SL ZXC21 and MERS-CoV. Phylogenetic analysis shows that Covid-19 is more closely related to Bat-SARS than to SARS-CoV [75]. Betacoronaviruses genome organization is given as figure 3 [78,79].
Betacoronaviruses genome organization, The Betacoronavirus for human genome comprises of the 50-Untranslated Region (50-UTR), open reading frame (orf) 1a/b (green box) encoding Non-Structural Proteins (nsp) for replication, structural proteins including spike (blue box), envelop (maroon box), membrane (pink box), and nucleocapsid (cyan box) proteins, accessory proteins (light gray boxes) such as orf 3, 6, 7a, 7b, 8 and 9b in the SARS-CoV-2 genome, and the 30-Untranslated Region (30-UTR). The doted underlined in red are the protein which shows key variation between SARS-CoV-2 and SARS-CoV.
There are five mutations which have been identified in Covid -19 including T9561C (codons TTA to TCA in ORF1a, a non-silent mutation), T8782C (codons AGT to AGC in ORF1a, a silent mutation), T29095C (codons TTT to TTC in Nucleocaspid, a silent mutation), C28144T (Codons TCA to TTA in ORF8b, a non-silent mutation), C15607T (codons CTA to TTA in ORF1b, a silent mutation [72].
S1 and S2 are the two subunits of spike glycoprotein, S1 subunit contains a single peptide followed by an N-Terminal Domain (NTD) and receptor-binding domain, while S2 subunit consists of four components such as conserved Fusion Peptide (FP), Heptad Repeat (HR) 1and 2, Transmembrane Domain (TM) and Cytoplasmic Domain (CP). S2 subunit of SARS-CoV-2 shows 99% identity with those of two bat SARS-like CoVs and human SARS-CoV whereas S1 subunit shows only 70% identity and S2 subunit is highly conserved. The development of broad spectrum antiviral peptides against this S2 subunit could be used for treatment modality for testing in animal models. The RBD component of S1 subunit have certain amino acid differences which is responsible for the direct interaction with the host receptor. Detailed studies on this soluble variable external subdomain region will give insight on pathogenesis, receptor usage and transmission of viruses among different species [73].
Identification, transmission and replication dynamics
Identification of Covid-19 using cell culture: When virus specimen was observed under the electron microscope, the spherical external spike protein displayss a characteristic crown shape. Samples were collected from the nasopharyneal and oropharyneal region of the patients. For the isolation of virus from oropharyneal samples, it was diluted with viral transfer medium containing nasopharyneal swabs and antibiotics and was incubated for 1 hour at 4ºC and then inoculated into Vero cells. After the inoculation, Vero cells were cultured in 1x Dulbecco's Modified Eagle´S Medium (DMEM) supplemented with 2% Fetal Bovine Serum (FBS) and penicillin-streptomycin, incubated at 37ºC and 5% CO₂. The cytopathic effects were observed from 3 days after inoculation, the inoculated cells were harvested on the 4th day when cytopathic effects in the cells were more than 80%. Virus isolation and replication were confirmed through different methodologies such as gene detection, cytopathic effects and electron microscopy. Virus replication was confirmed by RT-PCR using the RNA extracted from the cell culture medium, this RNA was amplified with primers for full- length gene analysis and NGS was performed using Miseq. According to the biosafety guidelines, all virus culture should be done inside Biosafety Cabinet Level-3 (bsl-3). Real-time RT PCR and transmission electron microscopy could be used for virus identification. In RT-PCR, the optimal concentration of primers and probes were required which were synthesized using known sequences and was determined with the RNA transcript of SARS-CoV. In transmission electron microscopy, the autolysis of the cells infected with virus could be prevented by inoculated cells which were prefixed by incubating with 2% paraformaldeyde and 2.5% glutaraldeyde in 0.1M phosphate buffer with pH 7.4. The virus could then be observed inside the cytoplasm of inoculated cells in electron microscope, virus particle size equal ranged from 70-90 nm. The aggregates of assembled intracellular virions could also be viewed inside the intracellular organelles, especially inside vesicles [80].
Transmission of Covid-19: Epidemiological studies revealed the fact that there are 3 factors involved in the transmission of nCoV-19 transmission which following:
- Source of infection: Natural host of SARS-CoV-2 were considered as Bats, from the study made by Wuhan institute of virology. They have found 96.2% similarity in the sequence between Covid-19 and bat coronavirus using gene sequencing technology. Pangolins were considered as intermediate host for Covid-19, studies made by technologies such as macrogenomic sequencing, molecular biological detection and electron microscopic analysis showed 99% similarity between Covid-19 isolated from pangolins and the virus strains currently infecting human beings [78].
- Route of transmission: The most common routes of transmission included droplets, aerosols and close contact with the infected patients. The respiratory droplets (size >5-10µm in diameter) produced by infected person during cough or sneeze is ingested or inhaled by nearby person which leads to transmission. The aerosols (having size <5um in diameter) are formed by mixing respiratory droplets into the air, when we inhale high dose of aerosols into the lungs, it lead to infection. Person after touching a surface or object contaminated with virus and subsequently touching other body parts such as nose, mouth and eyes will subsequently lead to contact based transmission of viruses [81]. Based on the evidence, reports suggest that digestive tract may be another route of infection since researchers have detected virus from stool, gastrointestinal tract, saliva and urine. The tears and conjunctival secretions of Covid-19 patients have also been found detected with presence of SARS-CoV-2 virus. More advanced studies are required to know more about the possibility of vertical transmission of virus between mother and infants [78]. Subclinical symptomatic or asymptomatic persons were considered as sources of Covid-19 infection. The virus invade CNS primarily through the olfactory route, yet grow better in primary human airway epithelial cells.
- Susceptibility of infection: Epidemiological studies report that older aged peoples are most susceptible to infection. The immunocompromised peoples like those with cancer, autoimmune disorders, AIDS, lung diseases, high blood pressure are also more vulnerable to infection. Based on clinical studies done, the median incubation period was 3 days (range 0-24) and the median time from symptom onset to death was 14 days, this median incubation period for Covid-19 was shorter than that for SARS and MERS infection. The maximum latency of Covid-19 were observed as 24 days, which might increase the risk of transmission. The disease progression rate in elderly people was also more rapid as compared with younger people [78].
Replication Dynamics of Covid-19: From the 16 Non-Structural Proteins (nsp) known, most of nsp's have specific role in replication. The nsp1 helps in cellular mRNA degradation by inhibiting IFN signaling, nsp3 have role in polypeptide cleaving thereby blocking host innate immune response and it also promotes cytokine expression, nsp4 helps in DMV formation, nsp6 helps in restricting autophagosome expansion and DMV formation, nsp9 have role in dimerization and RNA binding and nsp16 functions negatively regulating innate immunity [76].
Four structural proteins such as S proteins, M proteins, E protein and N protein play a major role in virion assembly and infection of CoVs. Homotrimers of S protein make up the spikes and these spikes on viral surface helps in attachment to host receptors. The structure of M protein contains 3 transmembrane domain, this M protein shapes the virion, promotes membrane curvature and binds to nucleocaspid. The E protein helps in virus assembly and release thereby involved in viral pathogenesis. The N protein helps to tether the genome to RTC and package the encapsidated genome into virions. The N protein play major role in viral replication by, act as antagonist of interferon and viral encoded repressor of RNA interference.
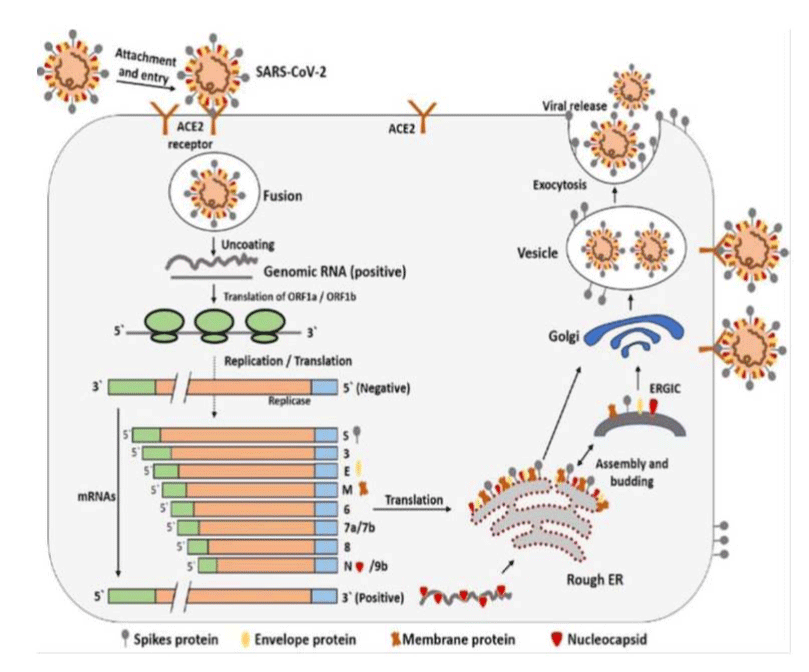
Human lower respiratory tract found ACE2 act as cell receptor for Covid-19 and regulates transmission of virus. The studies on Bronchoalveolar Lavae Fluid (BALF) isolated from Covid-19 infected patients, confirmed that Covid-19 and SARS-CoV uses the same cellular entry receptor, ACE2. The cellular receptor ACE2 found on the surface of human cells were used as a site of attachment for virion S-glycoprotein on the surface of coronavirus. In order to perform cell entry process, this spike protein need to be primed by protease enzyme called TMPRSS2 [79]. S1 and S2 are the two subunits of virion S-glycoprotein, both subunits perform different functions. S1 subunit determines the virus-host range and cellular tropism with RBD as key function domain, while S2 subunits helps in virus cell membrane fusion by two tandem domains such as Heptad Repeats 1 (HR1) and Heptad Repeats 2 (HR2). After the attachment of nCoV to the host cell membrane, membrane fuses and releases viral genome RNA into the cytoplasm and the uncoated RNA translates into two polyproteins such as pp1a and pp1ab, these polyproteins encode non-structural proteins and form RTC complex (replication-transcription complex) inside double-membrane vesicle. RTC complex replicate continuously and synthesize a nested set of subgenomic RNAs, which encodes structural proteins and accessory proteins, this mediates endoplasmic recticulum and golgi. The nucleocaspid proteins, envelop, glycoproteins and newly formed genomic RNA assemble together and forms viral particle buds. The nCoV genome replication and transcription takes place at cytoplasmic membrane of host cell. Finally virions will be released from the host cell by virion-containing vesicles which fuse with the plasma membrane of the host cell [74]. The life cycle of SARS-CoV-2 in host cells is given as figure 4 [79].
The life cycle of SARS-CoV-2 in host cells, after receptor binding, the conformation change in the S protein facilitates viral envelope fusion with the cell membrane through the endosomal pathway, then RNA into the host cell. Genome RNA is translated into viral replicase polyproteins, then cleaved into small products by viral proteinases. The polymerase produces a series of subgenomic mRNAs by discontinuous transcription and finally translated into relevant viral proteins. Viral proteins and genome RNA are subsequently assembled into virions and released out of the cell.
Epidemiological aspects of Covid-19: According to WHO situation reports as on May1 2020, there have been 31,75,207 confirmed cases of Covid-19 and 2,24,17 deaths due to Covid-19 globally [82]. This began as an outbreak in December 2019 in Wuhan, China [83] and on January 7, 2020, the causative pathogen was identified as a novel coronavirus which was different from known SARS-CoV and MERS-CoV, but closely related [84]. In earlier studies reported 49-66% patients had the contact history of Huanan seafood market, where different kinds of living wild animals were on sale, including poultry, bats, and marmots [26]. The environmental samples taken from Huanan seafood market were tested positive for SARS-CoV-2, but the exact animals associated with the virus have not been identified. Based on previous evidence, the bats, the host of more than 30 coronaviruses, may be the origin of Covid-19. In the initial phase, from the first case in December 2019 to the emergence of new cases outside Wuhan by January 13, 2020, a total of 41 cases were confirmed. Epidemiologic analysis showed that already in this initial phase, person-to-person transmission had occurred by close contact [85]. In the second phase which was in early and mid-January 2020, the virus spread to other Chinese provinces, On 20 January, China reported nearly 140 new cases in one day. Already by January 23, twenty nine provinces, and six foreign countries, had reported a total of 846 confirmed cases, which was about 20-fold increase from the first phase [86]. In the time of third phase which was by the end of January, the number increased 240-fold, reaching 9826 confirmed cases, and WHO declared this to be an epidemic and a Public Health Emergency of International Concern (PHEIC) [87]. According to the epidemiological analysis carried out by Chinese Center for Disease Control and Prevention, in first 72,314 cases of Covid-19 found in China, it was found out that this disease was highly contagious. It had spread exponentially from a single city to the entire country within only about 30 days. But according to the studies it was fortunate that Covid-19 had been mild for 81% of patients and had a very low overall case death rate of 2.3% in which a greater part have been ≥60 years of age or have had other pre-existing, conditions such as hypertension, cardiovascular disease, and diabetes [84].
By the end of January, the virus spread to all provinces of mainland China. By this time Covid-19 confirmed cases were reported in other countries such as South Korea, Iran, Italy, Spain, France, Germany, USA and India. As of 2 May 2020, more than 3.34 million cases of Covid-19 had been reported in 187 countries and territories, resulting in more than 2,38,000 deaths. More than 1.05 million people have recovered. United States of America has the most number of Covid-19 confirmed cases in the world followed by Spain, Italy, United Kingdom and Germany.
Pathogenesis
SARS-CoV-2 is spread mainly via respiratory droplet, contact, and potential in fecal-oral route. The infection of SARS-CoV-2 is of clustering onset, is more chances to infect older men with other diseases, and can result in severe and even fatal respiratory diseases such as Acute Respiratory Distress Syndrome [88]. The primary replication of SARS-CoV-2 arise in the mucosal epithelium of upper respiratory tract followed by further replication in lower respiratory tract and gastrointestinal mucosa. This may give rise to mild viremia. In some patients, the virus infection may be controlled at this stage and they remain asymptomatic SARS CoV-2 infect the human cells by spike glycoprotein binding to its cellular receptor, Angiotensin Converting Enzyme 2 (ACE2) [89,90]. This rapid viral replication in lower respiratory tract leads to cellular damage, virus-induced ACE2 downregulation and shedding, and Antibody Dependent Enhancement (ADE). ACE2 is widely expressed in nasal mucosa, bronchus, lungs, heart, esophagus, kidney, stomach, bladder, and ileum, which means all of these organs can be affected by SARS-CoV-2 [91]. This pathogen after reaching lower respiratory tract and gastrointestinal mucosa also induces over activation of T cells in peripheral blood, manifested by increase of Th17 and high cytotoxicity of CD8 T cells, leading to the severe immune dysfunction in the patient [92]. These all leads to cytokine storm ie, the production of exuberant pro-inflammatory cytokines and chemokines. ACE2 downregulation and shedding could also enhance inflammation and cause vascular permeability and pulmonary edema. This cytokine storm and pulmonary edema could be the major cause for ARDS, and even sudden death, though most patients survive the inflammatory responses and eliminate the virus.
According to the Single-cell RNA-seq data studies, expression of ACE2 might cause viral replication in organs such as lungs, liver kidney heart. The clinical symptoms such as dyspnea, diarrhea, acute cardiac injury, and kidney failure in Covid-19 patients might be due to this attack of the pathogen in the lung, upper respiratory track, ileum, heart, and kidney. The viral replication in these organs could lead to systemic viral sepsis followed by systemic inflammatory responses which leads to multiple organ damage and then death [90,91].
Virus host interactions
Initially due to the limited knowledge of SARS-CoV-2, the interaction of the pathogen to human cell is presumed to be similar to that of SARS-CoV. Later studies proved that the assumption was correct. The spike glycoprotein protein present on the surface of the virion, binds to the human Angiotensin-Converting Enzyme-2 (ACE2) membrane receptor thereby promoting its fusion to the host cell membrane. The spike protein of SARS-CoV-2, it contains two regions, S1 subunit and S2 subunit, which consists of 1253 amino acids. S1 domain is associated to receptor binding; S2 domain is associated to cell membrane fusion. Similar to SARS-CoV, S1 contains the N-Terminal Domain (NTD) and a Receptor-Binding Domain (RBD). S2 contains three functional domains, Fusion Peptide (FP), and Heptad Repeat (HR) 1 and 2. The RBD binds to the ACE receptor, which triggers a conformational change in the S protein, which then promotes membrane fusion between the viral and cell membrane through the S2 domain. The S2 changes conformation to facilitate the membrane fusion by three functional domains [89,93]. According to later studies it was found that the ACE2 binding affinity of the SARS-CoV 2 spike protein was 10−20-fold higher than that of the SARS-CoV spike protein [94]. Studies found out that the functional domains of S2 subunit, S-HR1 and S-HR2 play key roles in mediating SARS-CoV 2 fusion with and entry into the host cell. When S1 protein recognizes its receptor on human cells, the HR1 and HR2 domains are exposed to interact with each other, forming 6-HB to mediate membrane fusion between virus and target cell [95].
Impact of weather conditions
It has been already reported earlier that there are associations between climatic conditions and infectious diseases [96]. Certain environmental conditions can boost transmission of viruses. Not only temperature, but it is correlated to other factors such as relative humidity, or human behaviours during cold or warm weather. When the first case of corona virus was reported, the port city of Wuhan in China was experiencing its winter season, where the average temperatures ranged from 1 to 11 degrees Celsius [26]. The reason why cold weather could have been a cause in spreading of coughs, colds and flu is that cold air causes irritation in the nasal passages and airways, which makes people more susceptible to viral infections. Winter weather also tends to make people spend more time indoors which increases the risk of spreading the infection.
The cold weather reduces human's innate immunity. Cold temperature reduces the blood supply and thus reduces the provision of immune cells to the nasal mucosa [97,98]. According to disease experts, when the air is cold and dry, the droplets from sneeze or cough are more likely to float in the air for longer and travelling further and infecting more people [99].
Covid-19 pandemic might be partially suppressed with increasing temperature and humidity [100]. In the northern hemisphere, the warmer temperature might modestly reduce rate of spread of the virus, but a substantial decline in total number infected due to warmer temperatures could not be anticipated [97]. A study by Chin et al suggested that the Corona virus was highly stable at 4°C and sensitive to heat. The virus survival time had shortened to 5 min as the incubation temperature increased to 70°C [101]. Variation in transmission and migration rates could result in substantial variation in prevalence between regions. While the uncertainty in parameters is large, the scenarios shows that transient reductions in the incidence rate might be due to a combination of seasonal variation and infection control efforts and do not necessarily mean the epidemic is contained [98].
Quarantine during the COVID-19 pandemic
Quarantine could play a role in controlling the spread of coronavirus. Early implementation of quarantine along with other public health measures such as isolation, physical distancing would certainly reduce spread of the disease. Transmission between humans and secondary infections among close contacts and health-care workers, should be limited using strong quarantine protocols thus preventing transmission amplification events and preventing further international spread.
In line with the International Health Regulations, most of the countries around the world, had issued travel advisories from time to time, considering the surge in cases of Covid-19 in China. People travelling abroad was quarantined upon return. With rapid world wide spread of the virus, these travel restrictions had extended to almost all countries around the globe. At the community level, people are asked to avoid crowded areas and postpone non-essential travel to places with ongoing transmission. Diamond Princess, a cruise ship docked off Yokohama in Japan, was quarantined for two weeks after a tourist who disembarked at Hong Kong after testing people positive for Covid-19 [103,104]. If the pandemic threats are to be contained with lower magnitudes of loss to human life and economy, every country needs to invest in building up people-centric health systems, which pre-empt and prevent, rather than work in reactive, feedback loops driven by the burden of human misery [105]. In global setting, locking down Wuhan city was one of the immediate measure taken by Chinese authorities and hence had slowed the global spread of Covid-19 [106].
Being a new disease, there is only a limited information regarding risk factors for severe illness associated with Covid-19. The highest risk associated with corona virus infection is seen in adults ≥60 years of age, and in those with certain underlying conditions, such as cardiovascular and cerebrovascular diseases and diabetes [107,108]. Moderate-to-severe asthma, chronic lung diseases, Diabetes, Serious heart conditions, Chronic Kidney diseases and severe obesity, might also put people at higher risk for severe illness from Covid-19 [109]. Groups of persons at higher risk of infection and severe disease may require additional surveillance or they may require specific medical treatments. The communal nature of nursing homes and long-term care facilities put those living in nursing homes at higher risk of infection and severe illness from Covid-19.
Health care workers account for a significant proportion of infections in Corona virus outbreaks. Exposed Health care workers may experience a high incidence of infections, particularly for unprotected and repeated exposures [110]. Another risk factor in Health care workers include use of PPE kits [111]. Job that dealing with palm civets was the main risk factor of SARS-CoV infection in animal market workers [112], people who are immune-compromised, including those who are undergoing cancer treatment, bone marrow or organ transplantation, immune deficiencies, poorly controlled HIV or AIDS, and prolonged use of corticosteroids and other immune weakening medications all add to the risk group for acquiring the infection. Social distancing, frequent, thorough handwashing and other guidelines such as wearing a cloth face covering if social distancing isn't possible would all be appropriate to help lower the risk for everyone, especially the most vulnerable.
Vaccine development strategy
Every year, vaccines are administered to millions of people to mitigate the spread of a wide variety of infectious diseases. One of the best known and widely used vaccines is the influenza ("flu") vaccine. Prior to the widespread adoption of the flu vaccine, the influenza virus was associated with relatively high mortality rates. Acute viral respiratory tract infections remain a leading cause of morbidity, mortality, and economic loss. Although it doesn't affect much of younger and healthy adults, these infections are responsible for a substantial loss of productive time and are important contributing factors to the illness and death of the very young, of immunocompromised individuals, and of elderly populations.
Vaccine development often takes many years, several vaccine candidates are already moving out of the initial exploratory stage into the preclinical and clinical stages. Vaccine development comprises of several stages like exploratory stage, Preclinical stage, Clinical development, Regulatory review, approval, Manufacturing and Quality control. Several efforts were made to develop a vaccine against human coronavirus (CoV) infections such as MERS and SARS as well as for Covid-19. Many efforts have been made for developing CoV vaccines and drugs that target the spike glycoprotein or S protein, the major inducer of neutralizing antibodies. Although a few candidates have shown efficacy in In vitro studies, not many have progressed to randomized animal or human trials, hence may have limited use to counter Covid-19 infection [113].
Multiple strategies are adopted in the development of CoV vaccines; In November 2002, an outbreak of atypical pneumonia was reported in Guangdong Province, Southern China and on 12th March 2005, WHO issued a global alert, warning travellers and others to be aware of the signs and symptoms of SARS-CoV (Severe Acute Respiratory Syndrome), as the causative agent of the outbreak [114,115,14]. Eight thousand ninety six cases of SARS-CoV infection were reported in twenty nine countries, with a total of seven hundred and seventy four deaths. Vaccines targeting several animal CoVs have been developed, and some have been demonstrated to be efficacious in preventing viral infections [116]. However, a phenomenon of enhanced disease following vaccination has been observed in cats upon infection with feline infectious peritonitis virus following previous infection, vaccination, or passive transfer of antibody [117].
Several S-protein-based strategies have been attempted for developing CoV vaccines, e.g., use of full-length S protein or S1-Receptor-Binding Domain (RBD) and expression in Virus-Like Particles (VLP), DNA, or viral vectors [118,119]. The S protein has a major role in the induction of protective immunity during infection with SARS-CoV by eliciting neutralizing-antibodies and T-cell responses. Both recombinant proteins that contain RBD and the recombinant vectors that encode RBD could be used for developing the effective SARS-CoV vaccines [120].
Many different types of vaccines are being developed as possible potential candidates. Inactive or live-attenuated viruses, Virus-Like Particle (VLP), viral vectors, protein-based, DNA-based, and mRNA-based vaccines are being developed and some are now entering animal studies for assessment of toxicology. mRNA vaccines development is a new technology and as yet none have been licensed for use. Vaccination and safety testing is usually performed in a manner compliant with good laboratory practice and usually takes 3-6 months to complete. As most of the population is naïve to Covid-19, a significant time-gap is expected between the availability of the vaccine and the population gaining immunity.
Inactivated whole-virus vaccines
The efficacy of inactivated whole-virus, SARS-CoV vaccines were evaluated in mice.
Live attenuated vaccines
To date, live attenuated vaccines for SARS-CoV have not been evaluated in pre-clinical models. However, experiments were being done to generate cDNAs encoding the genomes of CoVs, including SARS-CoV.
Subunit/expressed-protein vaccines
Bisht, et al. reported development of a subunit vaccine consisting of a soluble baculovirus-expressed N-terminal fragment of the S protein.
Vectored vaccines
Several experiments have been done and reported the preclinical evaluation of vaccines utilizing other viruses as vectors for SARS-CoV proteins, like a chimeric parainfluenza virus.
DNA vaccines
DNA vaccines have been demonstrated to elicit strong induction of immune responses to viral pathogens in pre-clinical models, specifically in mice; however, clinical data on DNA vaccines in human subjects are limited.
Recombinant adenovirus-based vaccine
This recombinant vaccine expressing MERS- Co- S protein was reported to induce systemic IgG, secretory IgA, and lung resident memory T-cell responses when administered intra-nasally into BALB/c mice and provide long-lasting neutralizing immunity to MERS spike pseudotyped virus, thereby suggesting that the vaccine may confer protection against MERS-CoV [121]. Furthermore, Rabies Virus (RV) as a viral vector as well as Gram Positive Enhancer Matrix (GEM) as a bacterial vector has been used to express MERS-CoV S protein.
The guidance to control Covid-19 might be based on existing measures for MERS and SARS, with some further precautions due to the unknown nature of this new CoV. The infected individuals who are hospitalized should be immediately be given supportive treatment such as mechanical ventilation, ICU admission, and supportive care. Therapy may be comprised of supplementary oxygen or high flow nasal cannula oxygen therapy to reduce breathing stress. Furthermore, RNA synthesis inhibitors (like 3TC, TDF), remdesivir, neuraminidase inhibitors, peptide (EK1), anti-inflammatory drugs, abidol, Chinese traditional medicine, such as Lianhuaqingwen and ShuFengJieDu Capsules, could be the promising COVID-19 treatments [122]. Scientists, researchers and various health agencies put high efforts to stop the rapid spread of covid -19 by designing appropriate vaccines and therapeutics worldwide.
The time required for drug discovery programs to develop, evaluate, and obtain approval for a new potent anti-Covid-19 agent could take more than 10 years. In the present scenario, the development of a new therapeutic agent for Covid-19 is not a feasible option with regard to available time. Another option is to repurpose broadly acting antiviral drugs used for other viral infections. Such drugs have the advantage of easy availability, known pharmacokinetic and pharmacodynamic properties, solubility, stability, side effects, and also well-established dosing regimens. Among the evaluated drugs, both remdesivir and chloroquine were found to be highly effective in controlling Covid-19 in vitro. Oral administration of neuraminidase inhibitors such as oseltamivir has been used as an empirical drug for Covid-19 suspected cases in China hospitals even though there is no evidence of its efficacy. Among the evaluated drugs, both Remdesivir and chloroquine were found to be highly effective in controlling Covid-19 In vitro [123]. The study also pointed out that the three nucleoside analogs such as Ribavirin, Penciclovir, and Favipiravir may not have significant in vivo antiviral effects against Covid-19 since higher concentrations were required to reduce the viral infection In vitro.
Achievements in the development of vaccines and therapeutic agents for SARS- and MERS-CoV as well as recent ongoing progress for Covid-19 will facilitate the development of effective vaccines and therapeutics against this emerging virus.
Many drugs have been recently undergoing clinical trials for the treatment for Covid-19 that includes a low-cost antimalarial drug chloroquine and its derivative Hydroxychloroquine (HCQ), along with several other antiviral drugs. Chloroquine and hydroxychloroquine, two medicines currently used for the treatment of malaria which is prevalent not in tropical countries and certain autoimmune diseases, are being investigated worldwide for their potential to treat coronavirus disease. However, efficacy in treating Covid-19 is yet to be shown in studies. But chloroquine and hydroxychloroquine are presently authorized to be used by only patients and healthcare professionals or as part of clinical trials or national emergency use programmes for the treatment of Covid-19. Hydroxychloroquine was observed to be more potent than chloroquine in inhibiting SARS-CoV-2 In vitro. Hydroxy chloroquine sulfate 400mg given twice daily for 1 day, followed by 200 mg twice daily for 4 more days is recommended to treat SARS-CoV-2 infection. There are several clinical trials on potential antiviral therapies taking place. The therapies could be divided into two categories depending on their target. One that acts directly on the coronavirus, either by inhibiting crucial viral enzyme responsible for genome replication or by blocking viral entry into human cells. The other is designed to modulate the human immune system, either by boosting the innate response, which has a particularly important role against viruses, or by inhibiting the inflammatory processes that cause lung injury. Most of these drugs were originally used for the treatment of other pathogens and were repurposed for the current Covid-19 trials.
Since Covid-19 has rapidly developed into a worldwide pandemic with a significant health and economic burden, hundreds of clinical studies have been registered with the intention of discovering effective treatments. Since 2005, the International Committee of Medical Journal Editors (ICMJE) had recommended that all clinical trials should be registered in publicly available domains before they could be considered for publication [124]. The introduction of this requirement and other initiatives to increase clinical trial transparency has contributed to an increasing number of trials being recorded in online registries, such as ClinicalTrials.gov and the International Clinical Trials Registry Platform of the WHO.
Scientists can't assume their vaccine design will just work; they have to test and test again. They have to recruit thousands of people to ensure the safety and usefulness of a vaccine. The process has six important phases listed below:
Vaccine design
Scientists has to study the biology of a pathogen and decide on how they will get the immune system to recognize it.
Animal studies
A newly developed vaccine will be tested in disease induced animal models to demonstrate how it works and to study its extreme adverse effects.
Clinical trials (Phase I): These represent the first tests in human beings and test the safety and dose.
Clinical trials (Phase II): This is a deeper analysis of how a drug or vaccine actually works biologically. It involves a larger group of patients and assesses the physiological responses and interactions with the treatment, like in a coronavirus trial, they may assess if a vaccine stimulates the immune system in a certain way.
Clinical trials (Phase III): The final phase III trials employs an even greater amount of people tested over a long period of time.
Regulatory approval: The final hurdle sees the regulatory agencies, like the US food and drug administration, the European Medicines Agency (EMA) and Australia's Therapeutic Goods and Administration (ATGA).
Current clinical trials: An overview
As of March 28, 202 clinical trials could be retrieved with the search term "COVID-19" in the database [125] of the U.S. National Library of Medicine. The number jumped to 388 as of April 8, 2020. Among these trials are: Seven studies involving human plasma; Eleven studies involving Traditional Chinese Medicine (TMC); Fourteen studies involving stem cells, mostly mesenchymal stem cells; Sixteen studies involving dietary complements, including vitamin C, and honey; Twenty seven studies dedicated to vaccines; Fifty two studies involving proteins, including commercially available monoclonal antibodies; Seventy studies involving antiviral drugs; More than hundred studies involving other small molecules.
Large clinical trials are under way to generate the robust data needed to establish the efficacy and safety of chloroquine and hydroxychloroquine in the treatment of Covid-19. EMA welcomes these trials, which will enable authorities to give reliable advice based on solid evidence to healthcare professionals and patients. Considering the urgency and the pressure healthcare systems face to save lives during the Covid-19 pandemic, some countries, including the USA and France, have put strict protocols in place to allow the experimental use of these two medicines, for example, in patients with severe forms of Covid-19.
Over 85% of the clinical trials for either the prevention and/or treatment of Covid-19 have been registered in China, which is not surprising given that the country saw the outbreak of the disease first. The first clinical trials were registered within 1 month of Covid-19 identification and rapidly expanded after that. Public health initiatives have thus far successfully curtailed the previously exponential growth of Covid-19 cases in China. This has reduced the number of potential participants for clinical trials in China and the registration of new clinical trials has been in decline since. The Covid-19 pandemic represents the gravest global public health threat seen after the 1918 influenza outbreak and has rapidly become a global healthcare emergency. Clinical trials need to produce high-quality data that could be used objectively to assess potentials therapies for both the treatment and prevention of this global emergency.
Several clinical trials of novel Coronavirus which are registered in the China Clinical Trial Registry (ChiCTR), provide data bases and information references for clinical treatment. Around 232 studies were registered in chiCTR and the overall number of registrations were increased mainly in the Hubei province in china. The statistics of Covid-19 clinical trials registered with ChiCTR as of February 24th, 2020 were collected and descriptive analysis of registration characteristics. CMT drugs with high research frequency are chloroquine, lopinavir/ritonavir, and I-IFN; BI was Cell therapy, plasma therapy, Thymosin, and M/P-AB. There were problems of unclear classification of research types and irregular registration behaviour. Also, within the studies researched, heterogeneity exists for various dimensions. Different study design characteristics have led to significant differences in some aspects of the clinical trial. Timely summary analysis could provide more treatment options and evidence for clinical practice. Finally, statistical high-frequency research drugs could provide more treatment options and evidence-based evidence for the clinical practice. The design of Covid-19 clinical trials should give priority to "timeliness". The trial sample size needs to consider the balance between clinical and statistical significance, and its estimated volume reflects the reliability and repeatability of the research results [126].
On January 13, 2020, the US National institutes of Health and Moderna's infectious disease research team has begun a phase I clinical trial in collaboration with National Institute of Allergy and Infectious Diseases (NIAID), the US National institutes of Health and KPWHRI. It is the first testing in humans of the mRNA-1273 vaccine and would look to enroll a total of 45 healthy adult volunteers aged between 18 and 55 years. On January 23, 2020 Moderna announced funding award from CEPI to accelerate development of Messenger RNA (mRNA) vaccine against novel coronavirus.
Use of plasma from patients who have recovered from Covid-19 is expected to possess the potential benefit of providing disease-specific neutralizing antibodies, before targeted therapies could be developed. During the Ebola outbreak in 2014, WHO advised the use of convalescent plasma or whole blood therapies. However, a nonrandomised comparative study in 84 patients with Ebola found no associated improvement in survival [127]. There are currently 12 registered trials to investigate convalescent plasma or immunoglobulins in Covid-19.
The Covid-19 pandemic represents the gravest global public health threat seen since the 1918 influenza outbreak and has rapidly become a global health emergency. Clinical trials need to produce high quality data that can be used to objectively assess potentials therapies for both the treatment and prevention of this global emergency.
To date, there is no specific antiviral drug, vaccine or specific treatment available for Covid-19. Isolation remains the most effective measure for the containment of infection. Suspected cases will be isolated in a single room, while the confirmed cases will be accommodated in the same room [128]. The treatment is symptomatic and supportive therapy is the treatment management method followed by the health professionals, which includes; administration of antipyretic and analgesic, maintenance of hydration [129], immunomodulating therapy, organ function support, Bronchoalveolar Lavage (BAL), blood purification, mechanical ventilation as respiratory support, Extracorporeal Membrane Oxygenation (ECMO) and administration of antibiotics in the case of secondary bacterial infections [128,130].
As per WHO guidelines, patients with mild Covid-19 infection are provided symptomatic treatment (such as antipyretics for fever) and monitoring; oxygen therapy or empirical neuraminidase inhibitor therapy is considered for patients with severe infection; patients with critical infection are provided with advanced oxygen therapy or ventilatory support [131]. Currently, no vaccination is available. But there are many ongoing clinical trials testing various potential antiviral drugs, convalescent plasma transfusion, and vaccines for the treatment of Covid-19.
Remdesivir (GS-5734), a broad spectrum antiviral nucleotide prodrug developed for Ebola virus has shown activity against SARS-CoV-2 In vitro and preclinical studies have shown its efficacy in inhibiting MERS-CoV and SARS-CoV infections [26,132]. Remdesivir inhibits replication through the premature termination of RNA transcription and is capable of affecting a wide range of RNA viruses [133]. Other antiviral drugs that have been used in treating Covid-19 infection are Oseltamivir, Umifenovir [133], Interferon-α (IFN-α), Lopinavir/Ritonavir, Chloroquine phosphate [133,134], Ribavirin, Arbidol, hydroxychloroquine in combination with azithromycin [134]. Though several drugs claim to be safe and efficient for Covid-19 treatment, only two drugs have been allowed by FDA; hydroxychloroquine sulfate and chloroquine phosphate [50]. Some research studies claimed that ribavirin and IFN-α have offered synergetic effect in early stage [129]. Also, preliminary results from the clinical study recommend that Covid-19 patients could be treated with hydroxychloroquine in combination with azithromycin to cure the infection and to limit the transmission of the virus [134].
Infection Prevention and Control (IPC) is a practical, evidence-based approach in preventing, patients and health workers from being affected by infections. Without effective IPC it is impossible to control the spread of the virus. Considering the lack of effective treatment and rapid transmission rate of the Covid-19 pandemic, different strategies should be implemented in both short-term and long-term aspects as a preventive measure to avoid its further spread. At this moment, the best prevention is to avoid the exposure to the virus. The main IPC measures that reduce the risk of exposure are: (i) to wash hands with soap and water for at least 20 seconds, (ii) to use a hand sanitizer with at least 60% alcohol content, (iii) to avoid touching eyes, nose and mouth with unwashed hands, (iv) to avoid close contact with sick person, (v) to avoid mass gatherings, (vi) to maintain distance, about 2 arm's length (or at least 1 metre) in particular with those with respiratory symptoms, (vii) to cover mouth and nose with a cloth face cover, (viii) to cover coughs and sneezes with a tissue, and (ix) to clean and disinfect frequently touched surfaces daily [135].
Public health measures to prevent or slow down the transmission of Covid-19 includes; case isolation, identification and follow-up of contacts, environmental disinfection, use of Personal Protective Equipment (PPE) such as disposable isolation gown with fluid resistant characteristics, disposable gloves with coverage over gown cuffs, disposable respirators like N95 masks and Filtering FacePiece 2 (FFP2) masks, eye protection with goggles, and if possible a face shield over goggles. Wearing a medical mask is one of the preventive measures to limit the spread of respiratory infections. But this measure alone is not enough and should be combined with hand hygiene or other IPC measures to prevent human-to-human transmission. WHO has issued detailed guidelines for the use of medical masks in communities, at home and at health care facilities in areas that are reported by Covid-19 infection [136]. According to this guideline, health care workers are recommended to wear a medical mask when patients with suspected or confirmed cases are admitted, and to use a certified respirator like N95 or FFP2 when engaged in aerosol-generating procedures. The guideline also advises on the use of masks for healthy people in community settings.
A delay in detection and response has been recorded in China, U.S, as well as in other major countries, thereby affecting the local health systems. So, early diagnosis and supportive treatments are required to limit the spread of infection. The most important measures for reducing infection spread at the community level rely on case detection, isolation, contact tracing of positive cases followed by quarantine for those exposed, social distancing and lockdown [137]. In a scenario of rapidly spreading infection, there is an urgent need for taking the right measures to control the Covid-19 pandemic and any future infectious diseases. From the present situation, it is very clear that most of the countries were unprepared to face this pandemic, with respect to medications, vaccines, PPEs etc. So, there is a need for highly specialized research centres for viral diseases, bacterial illness etc. under the umbrella of WHO along with contributions from worldwide to combat any future calamity.
In the current review, we summarize and comparatively analyze the emergence, diagnosis, immunoinformatics, pathogenicity etc. of the Covid-19 pandemic, the third virus of coronaviridae family that caused infection and disease to arise in the 21st century. Covid-19 seems to be nearly close to SARS with regards to the clinical basis and less lethal than MERS, but even with lesser severe clinical picture, it could spread in the community more easily which have been presently reported in the nosocomial settings. In conclusion, there is still much more to unravel in the mystery of Covid-19, especially pertaining to its epidemiological features such as mortality and capacity to spread in a pandemic level, challenges in diagnostics and the development of vaccine and drug. The preparation for future pandemics with clear understanding about the nature of the disease is very important. Development of rapid identification diagnostic kits with specificity, public health interventions like social distancing, development of diagnostics for the monitoring in real time spread of the disease should become standard protocols while encountering a new viral disease. Public health officials should prepare the public for realistic scenarios and greater compliance with traumatic mediations ranging from stay-at-home orders, economic costs and end-of-life care that will enhance our ability to conquer future pandemics faster and at lesser cost. At present, there is no specific standard treatment for Covid-19. Given the high rate of transmission of this virus between humans across the globe, giving it a pandemic nature. The transmission, replication, structure, epidemiology, pathogenicity, virus host reactions of the virus discussed in this review article, might be helpful to structure and identify strategies for the development of novel drugs and vaccines. We have also discussed various approaches for developing effective vaccines and therapeutic combinations to cope with this viral outbreak.
The authors thankfully acknowledge the support and guidance of the management of HLL Lifecare Ltd. for reviewing and undertaking Covid management activities towards supporting Government of India in its efforts.
- Catrin S, Zaid A, Niamh ON, Mehdi K, Ahmed K, Riaz A, et al. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int J Surg. 2020; 76: 71-76. DOI: 10.1016/j.ijsu.2020.02.034
- Li F. Structure, function and evolution of coronavirus spike proteins. Annu Rev Virol. 2016; 3: 237-261. DOI: 10.1146/annurev-virology-110615-042301
- Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV-a target for vaccine and therapeutic development. Nat Rev. 2009; 7: 226-236. DOI: 10.1038/nrmicro2090
- Zhou Y, Jiang S, Du L. Prospects for a MERS-CoV spike vaccine. Expert Rev Vaccines. 2018; 17: 677-686. DOI: 10.1080/14760584.2018.1506702
- Smith TF, Waterman MS. Identification of common molecular subsequences. Journal of molecular biology. 1981; 147: 195-197. DOI: 10.1016/0022-2836(81)90087-5.
- Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC bioinformatics. 2007; 8: 424. DOI: 10.1186/1471-2105-8-424.
- Dos S FR, Buhler S, Nunes JM, Bitarello BD, França GS, Meyer D, et al. HLA supertype variation across populations: new insights into the role of natural selection in the evolution of HLA-A and HLA-B polymorphisms. Immunogenetics. 2015; 67: 651-663. DOI: 10.1007/s00251-015-0875-9
- Bhasin M, Raghava GP. Prediction of CTL epitopes using QM, SVM and ANN techniques. Vaccine. 2004; 22: 3195-3204. DOI: 10.1016/j.vaccine.2004.02.005.
- Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: Improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. The Journal of Immunology. 2017; 199: 3360-3368. DOI: 10.4049/jimmunol.1700893
- Alsahafi AJ, Cheng AC. The epidemiology of Middle East respiratory syndrome coronavirus in the Kingdom of Saudi Arabia, 2012-2015. International Journal of Infectious Diseases. 2014; 45: 1-4. DOI: https://doi.org/10.1016/j.ijid.2016.02.004
- World Health Organization. Consensus document on the epidemiology of severe acute respiratory syndrome [SARS]. 2003. https://tinyurl.com/y4onf6v9
- Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, Van Amerongen G, Van Riel, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. The Lancet. 2003; 362: 263-270. DOI: 10.1097/01.idc.0000104903.16995.a8
- Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. The Lancet. 2003; 361: 1319-1325. DOI: 10.1016/s0140-6736(03)13077-2.
- Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proceedings of the National Academy of Sciences. 2005; 102:14040-1405. DOI: 10.1073/pnas.0506735102.
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005; 310: 676-679. DOI: 10.1126/science.1118391.
- Shi Z, Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus research. 2008; 133: 74-87. DOI: 10.1016/j.virusres.2007.03.012.
- Han HJ, Wen HL, Zhou CM, Chen FF, Luo LM, Liu JW, et al. Bats as reservoirs of severe emerging infectious diseases. Virus research. 2015; 205: 1-6. DOI: 10.1016/j.virusres.2015.05.006.
- Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research. 2020; 24: 91-98. DOI: https://doi.org/10.1016/j.jare.2020.03.005
- World Health Organization. Summary table of SARS cases by country, 1 November 2002-7 August 2003. Weekly Epidemiological Record. 2003; 78: 310-311. https://tinyurl.com/y4qyzfzu
- Sharif-Yakan A, Kanj SS. Emergence of MERS-CoV in the Middle East: origins, transmission, treatment, and perspectives. PLoS Pathog. 2014; 10: e1004457. DOI: 10.1371/journal.ppat.1004457
- World Health Organization. Managing epidemics: key facts about major deadly diseases. World Health Organization; 2018. https://tinyurl.com/y2g5o8st
- Al-Abdely HM, Midgley CM, Alkhamis AM, Abedi GR, Lu X, Binder AM, et al. Middle East respiratory syndrome coronavirus infection dynamics and antibody responses among clinically diverse patients, Saudi Arabia. Emerging infectious diseases. 2019; 25: 753. DOI: 10.3201/eid2504.181595
- Fehr AR, Channappanavar R, Perlman S. Middle East respiratory syndrome: Emergence of a pathogenic human coronavirus. Annual review of medicine. 2017; 68: 387-399. DOI: 10.1146/annurev-med-051215-031152
- Azhar EI, Hui DS, Memish ZA, Drosten C, Zumla A. The Middle East respiratory syndrome [MERS]. Infectious Disease Clinics. 2019; 33: 891-905. DOI: doi.org/10.1016/j.idc.2019.08.001
- Huang C, Wang Y, Li X, Lili Ren, Jianping Zhao, Yi Hu, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020; 395: 497-506. DOI: 10.1016/S0140-6736(20)30183-5
- Russell TW, Hellewell J, Jarvis CI, Van Zandvoort K, Abbott S, Ratnayake R, et al. CMMID COVID-19 working group. Estimating the infection and case fatality ratio for Coronavirus Disease [COVID-19] using age-adjusted data from the outbreak on the Diamond Princess Cruise ship, February 2020. Eurosurveillance. 2020; 25: 2000256. DOI: 10.2807/1560-7917.ES.2020.25.12.2000256.
- Chan JFW, Kok KH, Zhuc. Kelvin Kai-Wang To, Shuofeng Yuan, Kwok-Yung Yuen. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes and Infections 2020. 9; 1: 221-236. DOI: 10.1080/22221751.2020.1719902
- Lu R, Zhao X, Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020; 395: 565-574. https://tinyurl.com/y3ogn86x
- Sohrabi C, Alsafi Z, O'Neill N, Khan M, Kerwan A, Al-Jabir A, et al. World Health Organization declares global emergency: A review of the 2019 novel Coronavirus [COVID-19]. International Journal of Surgery. 2020. DOI: 10.1016/j.ijsu.2020.02.034
- Aslam S, Mehra MR. COVID-19: Yet another coronavirus challenge in transplantation. The Journal of Heart and Lung Transplantation. 2020; 39: 408-409. DOI: 10.1016/j.healun.2020.03.007
- Chu DK, Pan Y, Cheng SM, Hui KP, Krishnan P, Liu Y, et al. Molecular diagnosis of a novel coronavirus [2019-nCoV] causing an outbreak of pneumonia. Clinical chemistry. 2020; 66: 549-55. DOI: https://doi.org/10.1093/clinchem/hvaa029
- Holshue ML, DeBolt C, Lindquist S, Kathy H Lofy, John Wiesman, Hollianne, et al. First Case of 2019 Novel Coronavirus in the United States. The New England Journal of Medicine. 2020: 929-936. DOI: 10.1056/NEJMoa2001191.
- Hossain I, Khan MH, Rahman MS, Mullick AR, Aktaruzzaman MM. The epidemiological characteristics of an outbreak of 2019 novel Coronavirus Diseases [COVID-19] in Bangladesh: A descriptive study. Journal of Medical Science and Clinical Research. 2020; 8: 145-151. DOI: 10.3760/cma.j.issn.0254-6450.2020.02.003.
- Zhang R, Liu H, Li F, Zhang B, Liu Q, Xiangwen Li, et al. Transmission and epidemiological characteristics of Novel Coronavirus [2019-nCoV]-Infected Pneumonia [NCIP]: Preliminary evidence obtained in comparison with 2003-SARS. MedRxiv. 2020. DOI: 10.1101/2020.01.30.20019836
- Liu RZ, Li F, Zhang B, Liu Q, Li X, Luo L, et al. Transmission and epidemiological characteristics of Novel Coronavirus [2019-nCoV]-Infected Pneumonia [NCIP]: Preliminary evidence obtained in comparison with 2003-SARS. DOI: 10.1101/2020.01.30.20019836
- Baruah V, Bose S. Immunoinformatics‐aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019‐nCoV. Journal of medical virology. 2020; 92: 495-500. DOI: https://doi.org/10.1002/jmv.25698
- Wen J, Aston J, Liu X, Ying T. Effects of misleading media coverage on public health crisis: A case of the 2019 novel coronavirus outbreak in China. Anatolia. 2020; 31: 331-336. DOI: 10.1080/13032917.2020.1730621
- Khatri P, Singh S, Belani NK, Leng YY, Lohan R, Wei LY, et al. YouTube as source of information on 2019 novel coronavirus outbreak: a cross sectional study of English and Mandarin content. Travel medicine and infectious disease. 2020; 101636. DOI: 10.1016/j.tmaid.2020.101636.
- Wax RS, Christian MD. Practical recommendations for critical care and anesthesiology teams caring for novel Coronavirus [2019-nCoV] patients. Canadian Journal of Anesthesia/Journal canadien d'anesthésie. 2020; 1-9. DOI: 10.1007/s12630-020-01591-x
- Chen W, Huang Y. To protect health care workers better, to save more lives with COVID-19. Anesthesia and Analgesia. 2020. DOI: 10.1213/ANE.0000000000004834
- Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, et al. Coronavirus disease 2019 [COVID-19]: A perspective from China. Radiology. 2020; 296: E15-E25. DOI: 10.1148/radiol.2020200490.
- Hellewell J, Abbott S, Gimma A, Bosse NI, Jarvis CI, Russell TW, et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. The Lancet Global Health. 2020; 8: e488-e496. DOI: 10.1016/S2214-109X(20)30074-7.
- Wang X, Chen S, Tan F, Gao GF. Recent Trends and Challenges with COVID-19-Africa, April 4, 2020. China CDC Weekly. 2020; 2: 365-369. DOI: 10.46234/ccdcw2020.094
- Badrfam R, Zandifar A. Coronavirus disease 2019 in Iran: The need for more attention to primary health care. Public Health. 2020; 182: 187. DOI: 10.1016/j.puhe.2020.03.010
- Song P, Karako T. COVID-19: Real-time dissemination of scientific information to fight a public health emergency of international concern. Bioscience trends. 2020; 14: 1-2. DOI: 10.5582/bst.2020.01056
- Majumder MS, Mandl KD. Early in the epidemic: Impact of preprints on global discourse about COVID-19 transmissibility. The Lancet Global Health. 2020; 8: e627-630. DOI: https://doi.org/10.1016/S2214-109X(20)30113-3
- Pandey SC, Pande V, Sati D, Upreti S, Samant M. Vaccination strategies to combat novel corona virus SARS-CoV-2. Life Sciences. 2020; 256: 117956. DOI: 10.1016/j.lfs.2020.117956
- National Institutes of Health. NIH clinical trial of investigational vaccine for COVID-19 begins. https://tinyurl.com/wcyl27j
- Daughton CG. Natural experiment concept to accelerate the Re-purposing of existing therapeutics for Covid-19. Global Epidemiology. 2020; 2: 100026. DOI: https://doi.org/10.1016/j.gloepi.2020.100026
- Dawoud D. Emerging from the other end: Key measures for a successful COVID-19 lockdown exit strategy and the potential contribution of pharmacists. Research in Social and Administrative Pharmacy. 2020. DOI: 10.1016/j.sapharm.2020.05.011
- Dheda K, Jaumdally S, Davids M, Chang JW, Gina P, Pooran A, et al. Diagnosis of COVID-19: Considerations, controversies and challenges in South Africa. Wits Journal of Clinical Medicine. 2020; 2: 3-10. DOI: 10.18772/26180197.2020.v2nSIa1
- Zhao S, Zhuang Z, Cao P, Ran J, Gao D, Lou Y, et al. Quantifying the association between domestic travel and the exportation of novel Coronavirus [2019-nCoV] cases from Wuhan, China in 2020: A correlational analysis. Journal of travel medicine. 2020; 27. DOI: https://doi.org/10.1093/jtm/taaa022
- Bao L, Deng W, Gao H, Xiao C, Liu J, Xue J, et al. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. BioRxiv. 2020. DOI: https://doi.org/10.1101/2020.03.13.990226
- Patel A, Jernigan DB, 2019-nCoV CDC Response Team. Initial public health response and interim clinical guidance for the 2019 Novel Coronavirus Outbreak - United States, December 31, 2019-February 4, 2020. MMWR Morb Mortal Wkly Rep. 2020; 69: 140-146. https://tinyurl.com/yx6xp3xb
- Russell TW, Hellewell J, Jarvis CI, Van-Zandvoort K, Abbott S, Ratnayake R, et al. Estimating the infection and case fatality ratio for COVID-19 using age-adjusted data from the outbreak on the Diamond Princess cruise ship. medRxiv. 2020. DOI: 10.2807/1560-7917.ES.2020.25.12.2000256
- Wang W, Xu Y, Gao R. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020; 323: 1843-1844. DOI: 10.1001/jama.2020.3786
- To KK, Tsang OT, Leung WS. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect Dis. 2020; 20: 565-574. DOI: https://doi.org/10.1016/S1473-3099(20)30196-1
- Ong SW, Tan YK, Chia PY, Lee TH, Ng OT, Wong MS, et al. Air, surface environmental, and personal protective equipment contamination by severe acute Respiratory Syndrome Coronavirus 2 [SARS-CoV-2] from a symptomatic patient. JAMA. 2020; 323: 1610-1612. DOI:10.1001/jama.2020.3227.
- Yu F, Yan L, Wang N, Siyuan Y, Linghang W, Yunxia T et al. Quantitative Detection and Viral Load Analysis of SARS-CoV-2 in Infected Patients. Clin Infect Dis. 2020. 71: 793-798. DOI: 10.1093/cid/ciaa345.
- Bholane KP. Impact of corona outbreak on global economy. Purakala with ISSN 0971-2143 is an UGC CARE Journal. 2020; 31: 126-133.
- Han X, Cao Y, Jiang N, Chen Y, Alwalid O, Zhang X, et al. Novel Coronavirus Pneumonia [COVID-19] progression course in 17 discharged patients: Comparison of clinical and thin-section CT features during recovery. Clinical Infectious Diseases. 2020; 71: 723-731. DOI: https://doi.org/10.1093/cid/ciaa271
- Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, et al. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000; 13: 129-1241. DOI: 10.1016/s1074-7613(00)00014-5
- Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochemical and biophysical research communications. 2009; 388: 621-625. DOI: https://doi.org/10.1016/j.bbrc.2009.08.062
- Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nature Reviews Immunology. 2006; 6: 644-658. https://tinyurl.com/yydkrd33
- Brown J, Wang H, Hajishengallis GN, Martin M. TLR-signaling networks: An integration of adaptor molecules, kinases, and cross-talk. Journal of dental research. 2011; 90: 417-427. DOI: https://doi.org/10.1177/0022034510381264
- Lessler J, Reich NG, Brookmeyer R, Perl TM, Nelson KE, Cummings DA. Incubation periods of acute respiratory viral infections: a systematic review. Lancet Infect Dis. 2009; 9: 291-300. DOI: 10.1016/S1473-3099(09)70069-6.
- Shokri S, Mahmoudvand S, Taherkhani R, Farshadpour F. Modulation of the immune response by Middle East respiratory syndrome coronavirus. J Cell Physiol. 2019; 234: 2143-2151. DOI: 10.1002/jcp.27155.
- Kikkert M. Innate Immune Evasion by Human Respiratory RNA Viruses. J Innate Immun. 2020; 12: 4-20. DOI: https://doi.org/10.1159/000503030
- Faure E, Poissy J, Goffard A, Fournier C, Kipnis E, Titecat M, et al. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PLoS One. 2014; 9: 88716. https://tinyurl.com/yyy77k2a
- Jeannette Guamer. Three Emerging Coronavirusers in Two Decades: The Story of SARS, MERS and Now COVID-19. Am J Clin Pathol. 2020. 153: 420-421. DOI: 10.1093/ajcp/aqaa029
- Hanuman Singh Dagur, Saurabh Singh Dhakar. Genome Organization of Covid-19 and Emerging Severe Acute Respiratory Syndrome Covid-19 Outbreak: A Pandemic. EJMO. 2020; 4: 107-115. DOI: 10.14744/ejmo.2020.96781
- Jasper FWC, Kin HK, Zheng Z, Hin C, Kelvin KWT, Shuofeng Y, et al. Genomic characterization of the 2019 novel humanpathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes & Infections. 2020; 9: 221-236. DOI: 10.1080/22221751.2020.1719902.
- Yan RG, Qing DC, Zhon SH. Military medical research. 2020; 11: 1-3.
- Yu Chen, Qianyun Liu, Deyin Guo. Emerging coronaviruses: Genome structure, replication and pathogenesis. Journel of Medical Viroloy. 2020; 92: 418-423. DOI: 10.1002/jmv.25681
- Rozhgar A K, Muhamad S, Mehmet Ozaslan. Genomic characterization of a novel SARS-CoV-2. Elsevier Gene reports 19. 2020; 1-5. DOI: 10.1016/j.genrep.2020.100682
- Leila M, Sorayya G. Genotype and Phenotype of COVID-19: Their role in pathogenesis. Journal of Microbiology, Immunology and Infection. 2020; 1-4. DOI: https://doi.org/10.1016/j.jmii.2020.03.022
- Lisheng Wang, Yiru Wang, Dawei Ye. Review of the 2019 novel Coronavirus [SARS-CoV-2] based on current evidence. International journal of antimicrobial agents. 2020; 12: 2-3. DOI: https://doi.org/10.1016/j.ijantimicag.2020.105948
- Muhammad AS, Suliman K, Abeer K. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research. 2020; 24: 93. DOI: https://doi.org/10.1016/j.jare.2020.03.005
- Jeong MK, Yoon SC, Hye JJ, Mi SK, Sang HW, Sehee P, et al. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res Perspact. 2020; 11: 4-5. DOI: 10.24171/j.phrp.2020.11.1.02
- Sasmita PA, Sha M, Yu JW, Yu PM, Rui XY, Qing ZW, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of Coronavirus disease [COVID-19] during the early outbreak period: A scoping review. Infectious Diseases of poverty. 2020; 929: 8-9. DOI: 10.1186/s40249-020-00646-x
- World health organization, Coronavirus disease 2019 [COVID-19] Situation Report. 102. https://tinyurl.com/wl8kbzw
- Zhu N, Zhang D, Wang W, Li XW, Yang B, Song JD, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382: 727-733. DOI: 10.1056/NEJMoa2001017
- Surveillances, V. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases [COVID-19]-China, 2020. China CDC Weekly. 20202; 2: 113-122.
- Qun Li, Xuhua Guan, Peng Wu, Xiaoye Wang, Lei Zhou, Yeqing Tong, Ruiqi Ren, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med 2020; 382: 1199-1207. DOI: 10.1056/nejmoa2001316
- Sun J, He WT, Wang L, Lai A, Ji X, Zhai X, et al. COVID-19: Epidemiology, evolution, and cross-disciplinary perspectives. Trends in Molecular Medicine. 2020; 26: 483-495. DOI: https://doi.org/10.1016/j.molmed.2020.02.008
- Wang X, Zhang X, He J. Challenges to the system of reserve medical supplies for public health emergencies: Reflections on the outbreak of the severe acute Respiratory Syndrome Coronavirus 2 [SARS-CoV-2] epidemic in China. Bioscience trends. 2020; 14: 3-8. DOI: 10.5582/bst.2020.01043.
- Habibi R, Burci GL, de Campos TC, Chirwa D, Cina M, Dagron S, et al. Do not violate the International Health Regulations during the COVID-19 outbreak. The Lancet. 2020; 395: 664-666. DOI: https://doi.org/10.1016/S0140-6736(20)30373-1
- Ge H, Wang X, Yuan X, Xiao G, Wang C, Deng T, et al. The epidemiology and clinical information about COVID-19. European Journal of Clinical Microbiology & Infectious Diseases. 2020; 1-9. DOI: 10.1007/s10096-020-03874-z
- Jin Y, Yang H, Ji W, Wu W, Chen S, Zhang W, et al. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses. 2020; 12: 372. DOI: 10.3390/v12040372.
- Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Frontiers of medicine. 2020; 12: 1-8. DOI: 10.1007/s11684-020-0754-0
- Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet respiratory medicine. 2020; 8: 420-422. DOI: https://doi.org/10.1016/S2213-2600(20)30076-X
- Lim YX, Ng YL, Tam JP, Liu DX. Human coronaviruses: A review of virus–host interactions. Diseases. 2016; 4: 26. DOI: 10.3390/diseases4030026
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020; 367: 1260-1263. DOI: 10.1126/science.abb2507.
- Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cellular & Molecular immunology. 2020; 11: 1-3. DOI: 10.1038/s41423-020-0374-2
- Patz JA, Graczyk TK, Geller N, Vittor AY. Effects of environmental change on emerging parasitic diseases. International journal for parasitology. 2000; 30: 1395-1405. DOI: https://doi.org/10.1016/S0020-7519(00)00141-7
- Bannister-Tyrrell M, Meyer A, Faverjon C, Cameron A. Preliminary evidence that higher temperatures are associated with lower incidence of COVID-19, for cases reported globally up to 29th February 2020. MedRxiv. 2020.
- Neher RA, Dyrdak R, Druelle V, Hodcroft EB, Albert J. Potential impact of seasonal forcing on a SARS-CoV-2 pandemic. Swiss medical weekly. 2020; 150. DOI: https://doi.org/10.4414/smw.2020.20224
- Maboloc CR, Baratipour M, Parahakaran S, D'Astous M. Eubios Journal of Asian and International Bioethics. 2020; 30: 3.
- Wu Y, Jing W, Liu J, Ma Q, Yuan J, Wang Y, et al. Effects of temperature and humidity on the daily new cases and new deaths of COVID-19 in 166 countries. Science of the Total Environment. 2020; 729: 139051. DOI: https://doi.org/10.1016/j.scitotenv.2020.139051
- Chin A, Chu J, Perera M, Hui K, Yen HL, Chan M, et al. Stability of SARS-CoV-2 in different environmental conditions. medRxiv. 2020. DOI: https://doi.org/10.1101/2020.03.15.20036673
- Sun Z, Thilakavathy K, Kumar SS, He G, Liu SV. Potential factors influencing repeated SARS outbreaks in China. International Journal of Environmental Research and Public Health. 2020; 17: 1633. DOI: 10.3390/ijerph17051633.
- Rocklov J, Sjodin H, Wilder-Smith A. COVID-19 outbreak on the Diamond Princess cruise ship: Estimating the epidemic potential and effectiveness of public health countermeasures. Journal of travel medicine. 2020; 27: taaa030. DOI: 10.1093/jtm/taaa030.
- Dooley B, Rich M. Cruise ship's coronavirus outbreak leaves crew nowhere to hide. New York Times, 2020. https://tinyurl.com/tnbptn2
- Chatterjee P, Nagi N, Agarwal A, Das B, Banerjee S, Sarkar S, et al. The 2019 novel Coronavirus disease [COVID-19] pandemic: A review of the current evidence. Indian Journal of Medical Research. 2020; 151: 147. DOI: 10.4103/ijmr.IJMR_519_20.
- Heymann DL, Shindo N. COVID-19: What is next for public health? The Lancet. 2020; 395: 542-545. DOI: https://doi.org/10.1016/S0140-6736(20)30374-3
- Bai Y, Yao L, Wei T, Tian F, Jin DY, Chen L, et al. Presumed asymptomatic carrier transmission of COVID-19. Jama. 2020; 323: 1406-1407. DOI: 10.1001/jama.2020.2565
- Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020; 323: 1061-1069. DOI: 10.1001/jama.2020.1585
- Wu Z, McGoogan JM. Characteristics of and important lessons from the Coronavirus disease 2019 [COVID-19] outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. Jama. 2020; 323: 1239-1242. DOI: 10.1001/jama.2020.2648.
- Rando HM, Greene CS, Robson MP, Boca SM, Wellhausen N, Lordan R, et al. SARS-CoV-2 and COVID-19: An evolving review of diagnostics and therapeutics. Manubot. 2020.
- Leite P, Armada MR. Bond fund performance during recessions and expansions: Empirical evidence from a small market. International Review of Finance. 2017; 17: 163-1670. DOI: https://doi.org/10.1111/irfi.12098
- Wang M, Xu HF, Zhang ZB, Zou XZ, Gao Y, Liu XN, et al. Analysis on the risk factors of severe acute respiratory syndromes coronavirus infection in workers from animal markets. 2004; 25: 503. DOI: https://doi.org/10.1111/irfi.12098
- Kuldeep DA, Khan SB, Ruchi TC, Maryam DD, Yashpal SME, Karam PS, et al. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Human vaccines & immunotherapeutics. 2020; 1-7. DOI: 10.1080/21645515.2020.1735227
- Drosten C, Gunther S, Preiser W, Van Der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003; 348: 1967-1976. DOI: 10.1056/NEJMoa030747
- Keyaerts E, Vijgen L, Maes P, Neyts J, Van Ranst M. Growth kinetics of SARS-coronavirus in Vero E6 cells. Biochemical and biophysical research communications. 2005; 329: 1147-1151. DOI: 10.1016/j.bbrc.2005.02.085
- Saif LJ. Animal coronavirus vaccines: Lessons for SARS. Developments in biologicals. 2004; 119: 129-140. https://tinyurl.com/y48yv7m7
- Petersen NC, Boyle JF. Immunologic phenomena in the effusive form of feline infectious peritonitis. American Journal of Veterinary Research. 1980; 41: 868-876. https://tinyurl.com/yye44odk
- Graham RL, Donaldson EF, Baric RS. A decade after SARS: Strategies for controlling emerging coronaviruses. Nature Reviews Microbiology. 2013; 11: 836-848. https://tinyurl.com/y4je9c3j
- Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, et al. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004; 428: 561-564. DOI: 10.1038/nature02463.
- Jiang S, He Y, Liu S. SARS vaccine development. Emerging infectious diseases. 2005; 11: 1016. DOI: 10.3201/1107.050219.
- Gillim RL, Subbarao K. Emerging respiratory viruses: challenges and vaccine strategies. Clinical microbiology reviews. 2006; 19: 614-636. DOI: 10.1128/CMR.00005-06
- Wang Z, Chen X, Lu Y, Chen F, Zhang W. Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Bioscience trends. 2020; 14: 64-68. DOI: 10.5582/bst.2020.01030
- Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel Coronavirus [2019-nCoV] In vitro. Cell research. 2020; 30: 269-271. DOI: 10.1038/s41422-020-0282-0
- Smith R. Medical journals are an extension of the marketing arm of pharmaceutical companies. PLoS med. 2005; 2: e138. DOI: 10.1371/journal.pmed.0020138
- JJ VE. COVID-19: A Brief overview of the discovery clinical trial. Pharmaceuticals [Basel, Switzerland]. 2020; 13. DOI: 10.3390/ph13040065
- Abbas I. Hybrid research simulation modeling for making decisions on sample size and power of randomized clinical trials considering expected net benefits. In2017 Winter Simulation Conference [WSC]. IEEE. 2017; 2845-2856. https://tinyurl.com/y4wlxgof
- Van Griensven J, Edwards T, de Lamballerie X, Semple MG, Gallian P, Baize S, et al. Evaluation of convalescent plasma for Ebola virus disease in Guinea. New England Journal of Medicine. 2016; 374: 33-42. DOI: 10.1056/nejmoa1511812
- Chen ZM, Fu JF, Shu Q, Chen YH, Hua CZ, Li FB, et al. Diagnosis and treatment recommendations for pediatric respiratory infection caused by the 2019 novel coronavirus. World Journal of Pediatrics. 2020; 16: 1-7. DOI: 10.1007/s12519-020-00345-5.
- Kumar D, Malviya R, Sharma PK. Corona virus: A review of COVID-19. EJMO. 2020; 4: 8-25. DOI: 10.14744/ejmo.2020.22984
- Tang B, Xia F, Tang S, Bragazzi NL, Li Q, Sun X, et al. The effectiveness of quarantine and isolation determine the trend of the COVID-19 epidemics in the final phase of the current outbreak in China. International Journal of Infectious Diseases. 2020; 95: 288-293, DOI: https://doi.org/10.1016/j.ijid.2020.03.018
- Wang L, Li X, Chen H, Yan S, Li D, Li Y, Gong Z. Coronavirus disease 19 infection does not result in acute kidney injury: an analysis of 116 hospitalized patients from Wuhan, China. Am J Nephrol. 2020; 51: 343-348. DOI: https://doi.org/10.1159/000507471
- Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Science translational medicine. 2017; 9: eaal3653. DOI: 10.1126/scitranslmed.aal3653
- NaserGhandi A, Allameh SF, Saffarpour R. All about COVID-19 in brief. New Microbes and New Infections. 2020; 35. https://tinyurl.com/yyhcvey4
- Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 [COVID-19]. Drug discoveries & therapeutics. 2020; 14: 58-60. DOI: 10.5582/ddt.2020.01012
- Hassan SA, Sheikh FN, Jamal S, Ezeh JK, Akhtar A. Coronavirus [COVID-19]: A review of clinical features, diagnosis, and treatment. Cureus. 2020; 12: e7355. DOI:10.7759/cureus.7355
- World Health Organization. Rational use of personal protective equipment for Coronavirus disease [COVID-19]: interim guidance, 27 February 2020. World Health Organization; 2020. https://tinyurl.com/y3w6cmar
- Khanna RC, Cicinelli MV, Gilbert SS, Honavar SG, Murthy GS. COVID-19 pandemic: Lessons learned and future directions. Indian Journal of Ophthalmology. 2020; 68: 703-710. DOI: 10.4103/ijo.IJO_843_20.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.