> Medicine. 2020 June 22;1(2):039-047. doi: 10.37871/jels1118.
Antibiogram Pattern of Potential Pathogenic Bacteria Associated with Domestic Dog Faecal Matter in Port Harcourt Metropolis
Azuonwu Obioma1*, Ihua Nnenna1 and Ahiakwo Christian2
2Department of Animal and Environmental Biology, Rivers State University, Nkpolu-Oroworukwo, Port Harcourt, Nigeria
- Antibiogram pattern
- Potential pathogenic bacteria
- Domestic dog
- Faecal matter
- Zoonosis
- Resistant strains
This study was aimed at investigating the Antibiogram pattern of Potential Pathogenic Bacteria that are associated with Domestic dog faecal matter in Port Harcourt Metropolis. Convenient sampling techniques were explored for sample collection. A total of fifty samples were collected from domestic dogs from different locations (Agip estates, Rumuokoro, GRA, Sandfill, Town, RSU lecturers’ quarters) within Port Harcourt metropolis, Rivers State, Nigeria. The bacteriological analysis was determined using standard microbiological procedures and identification techniques. Potential pathogens associated with domestic dog faecal matter that were isolated include; Escherichia coli, Klebsiella species, Pseudomonas species, Proteus species, Bacillus species, Staphylococcus aureus and Streptococcus species. However other species of Staphylococcus were also isolated respectively. The study showed that among the seven antibiotics used, tarivid was the most sensitive (96%), while peflacin was the most resistant (62%). The correlation analysis showed the relationship between isolates and bacterial count with antibiogram revealed that only Ciproflaxacin showed significant correlation with the isolates (r = 0.322, p = 0.02). This study strongly suggests that domestic dogs carry potential pathogenic organisms present in the faecal matter that can serve as sources of infection to the pet owners, especially the strains showing resistance to antibiotics. These pose a community health threat, thus putting the general public at risk of contracting infections. It is, therefore, important that these domestic dogs including the ones used as pets should be treated and possibly vaccinated frequently, even as faecal matter from domestic dogs should be well disposed to prevent possible zoonotic infections to man from the contaminated environment.
Background
The increase in the population of stray domestic and semi-domestic dogs in urban areas across the globe has increased the risk of zoonotic diseases. Domestic dogs are either kept for security reason or as pets assets most times. World Health Organization (2018) reported that about 5 million people throughout the world are annually bitten by dogs [1]. Many parasitic and zoonotic pathogens are transmitted by dogs through this obvious process [2]. Nonetheless, enormous numbers of diseases that affect humans today originated from animals otherwise known as zoonotic infection. Although pets come with many benefits, as observed within the communities across the world, however, pets as a matter of clinical importance occasionally carry harmful germs that can make people sick, if precautionary measures are not put in place in good time. Nevertheless, the diseases people get from animals are called zoonotic diseases and it is difficult and highly complicated to know which animals could be carrying zoonotic diseases, especially since most animals carrying these germs often look healthy and normal, in the obvious scenario, asymptomatic carriers even when they are shedding the pathogens within their environment and living space [3]. Hence, some potential pathogenic bacteria have been shown to be associated with domestic dogs in our communities or hinterlands.
Zoonotic diseases of dogs are caused by a range of agents from virus, parasites, fungi and bacteria origin. The most common zoonotic diseases of dogs are ringworm, salmonellosis, leptospirosis, Lyme disease, campylobacter infection, Giardia infection, cryptosporidium infection, roundworms, hookworms, tapeworms, scabies, harvest mites and rabies. Diseases of the gastrointestinal tract characterized diarrhoea as one of the most common clinical manifestations, potentially leading to severe dehydration, and zoonosis has been implicated as one of the causes of death [4]. Notably, this was reported by previous studies like Salmonellosis and other gastro-enteritis [4-6].
Nevertheless, domestic dogs particularly Pets offer comfort and companionship, and one cannot help but love them. While pets can benefit our health and emotional wellbeing in several ways, they have also had been implicated as a potential source of the spread of infections, and has caused various human illnesses in the world undoubtedly [7]. Dog faeces constitute public health nuisance, thus in a study carried out on dog faeces in Italy, microorganisms such as Salmonella, Yersinia or Campylobacter species, Giardia cysts, Enterococcus species, E. faecium, E. gallinarum, E. casseliflavus., E. faecalis were isolated [8]. These could be potential sources of environmental contamination to man and other vulnerable creatures within the ecosystem.
Nonetheless, antibiogram pattern of potential pathogenic bacteria associated with domestic dog demonstrates a high degree of variance in the different geographical landscape as seen from different studies outside and within the region of this present study. However, some studies maintained similar resistance and susceptibility rate and pattern. Nevertheless, it is strongly believed that since the early 20s, there has been an increased indication that pets and faecal matter from pets may probably be a reservoir for antibiotic-resistant bacteria strains of dangerous perspective, thus, posing a new threat to public health intervention programs and strategies across the global communities. Some strains have been found to be resistant to some antibiotics including clindamycin, vancomycin, tetracycline, erythromycin and ampicillin at varying degrees [9,10]. Thus, these should be worrisome to microbiologist, clinicians and other numerous stakeholders in the medical profession given the overall medical implication of such outcome in our developing communities with weak, and fragile health facilities to handle such complicated and non-cost-effective treatment options among the poor citizens.
However, it is strongly reported that a comparatively high occurrence (7-23%) of Vancomycin-resistant enterococcus, mainly Enterococcus faecium in dogs living in urban areas has also been reported in Europe [11]. Moreover, enterococci with High-Level Aminoglycoside Resistance (HLAR) have been described in strains isolated from both humans and animals [12]. In addition, Methicillin-Resistant Staphylococcus aureus (MRSA) has been found in the faecal matters of dogs and has been isolated from both infected and colonised pet animals [13]. The MRSA isolated from pet animals resembles MRSA strains isolated in a hospital setting suggesting transmission and cross infection of these strains from animals to human or vice versa [14]. Of additional concern is also the description of the isolation of Escherichia coli producing extended-spectrum beta-lactamases from dogs [15]. Thus, dogs represent a potential source of antimicrobial-resistant microorganisms, particularly considering the overuse of antimicrobials in companion animals. Salmonella species are anaerobic and motile gram-negative bacilli that colonize the large intestine of a variety of mammals, especially in the distal part of the colon and the mesenteric lymph nodes of the canine. Humans can also get infected through the gastrointestinal tract (faecal transmission) and cultivate several infectious diseases such as gastroenteritis, enteric fever, bacteraemia and osteomyelitis. Gastrointestinal diseases are the most prevalent clinical presentations of salmonella in human and dogs; nevertheless, the majority of infected animals or humans are asymptomatic and may shed the pathogen through faeces for a period of 6 weeks and transmit the same pathogen to other animals or individuals within their vicinity. In developing nations, Salmonella species are also more prevalent than in developed countries [16]. An antibiogram laboratory assay should be considered for patients infected with Salmonella species however, it could be treated by various families of antibiotics including fluoroquinolones, beta-lactams, and macrolides but conducting the analysis first remains critical for specificity and sensitivity of the overall outcome [17].
There are Risk Factors and Public Health implication of potential pathogenic bacteria that are associated with domestic dogs. Putative risk factors for acute infectious diarrhoea in canines include breed, gender, vaccination history, age, season, breeder-origin, and/or stay in kennel [18]. However, these risk factors for infection with specific enteropathogens have mainly been reported in dogs housed in shelter or breeding facilities. The importance of some of these risk factors might be different for privately owned dogs kept under different living conditions. This, however, is not within the scope of this study for record purposes.
Nonetheless, Africa and Asia have the highest global burden of Rabies, a zoonotic disease commonly encountered through bites from dogs. This probably expresses the ineffectiveness of the eradication program in Nigeria. Studies revealed that exposure to rabies through bites from rabid dog accounts for 100% of the confirmed cases in Nigeria [19-21]. In Nigeria, there is an existing dog anti-rabies vaccination schedule at twelve weeks of age [22]. Nigeria has a guideline for veterinary and pet control but the rate of non-compliance may probably account for high cases of Rabies in the country compared to some countries like United Kingdom, New Zealand, Hawaii, Australia, Japan and Antarctica which are free from the rabies virus through successful eradication programs [23].
However, the dearth of data with respect to the above subject matter stimulated the reason and passion for the study. Furthermore, coupled with the varying degree of conclusions from the existing few studies encouraged the researchers to consider this direction of study, particularly with the increasing demands of dogs as pets and for security use world-wide. Nevertheless, the focus and the objectives that guided this study are as follows:
(1) To identify some Potential Pathogenic Bacteria that are Associated with Domestic Dog Stool in Port Harcourt Metropolis.
(2) To determine the Prevalence of Antibiotics resistance and susceptibility of the Potential Pathogenic Bacteria that are Associated with Domestic Dog Stool.
(3) To compare the Mean difference of the minimum zone of inhibition (antibiogram) of the various Potential Pathogenic Bacteria that are Associated with Domestic Dog Stool in Port Harcourt Metropolis
(4) To examine the correlation between Antibiogram pattern and Potential Pathogenic Bacteria isolated from Domestic Dog Stool as well as with the bacteria count.
Interestingly, it is strongly believed that data generated would enhance the curiosity of government and her relevant agencies to strengthen the policies of care for dogs and the need for compulsory vaccination of all domestics animal pet, to reduce the trend of zoonotic infections and multidrug resistance menace in our society at large, even as this study would also serve as a baseline study for further studies in this direction in the region, given the scarcity of data of similar studies in the region.
Sample area
Samples for this study were collected from six (6) different locations in Port Harcourt (Agip estates, Rumuokoro, GRA, Sandfill, Town and RSU lecturers’ quarters). Port Harcourt, Port town and capital of Rivers State, Southern Nigeria. It lies along the Bonny River (an eastern distributary of the Niger River) 41 miles (66 km) upstream from the Gulf of Guinea and is located in the Niger Delta [24]
Sample size derivation
where Z is the statistics corresponding to the level of 95% confidence of 1.96
P = expected prevalence in percentage (from similar study) = 0.0345 [25] d = level of significance (above error) of 5%
5% = 0.05
1-p = 1-0.0345 = 0.9655
Sample collection/experimental
The dog faecal samples were collected aseptically into the sterile bottles and transferred immediately to the laboratory in an ice parked package for microbiological culture and processing technique. The samples were collected from 50 pet dogs from different locations and were cultured on blood, chocolate Mac Conkey, Salmonella-Shigella, Deoxycholate citrate and Nutrient agar respectively. The spreading of the sample on a plated media was done to achieve the heterotrophic plate count outcome, hence a serial dilution of 1:10 was explored, and the already sterilized glass rod spreader was employed to spread the first primary inoculum in duplicates, after it was dropped with a sterilized wire loop, then streaking follows with a sterilized wire loop to produce discrete colonies after 24 hours of incubation at a temperature of 37°C. The organisms were subcultured and isolated to obtain a pure culture accordingly as described by Cheesbrough [26].
Identification of the isolated pathogens
Also, Gram staining and microscopy examination technique were explored for the identification of the pathogens. Furthermore, biochemical test analysis such as catalase, citrate, coagulase, indole, oxidase and, mannitol salt agar test assay were used to further underpin the pathogen identification as described by Cheesbrough [2006].
Susceptibility antimicrobial assay
The Kirby-Bauer disc method as described by Cheesbrough [26] was explored for the antimicrobial susceptibility assay. The selected isolated organisms were aseptically emulsified after mixing properly in a sterile saline solution to ensure that no solid particles could be seen visible in a saline solution. The process was continued until the turbidity of the solution was virtually matched with that of the standard turbidity reference outcome. A representative sample of the organism for the assay in the tube was picked and was used to streak completely on Mueller-Hinton agar plate, with the help of standard wire loop, an antibiotic disc were aseptically placed on the surface of the agar, after it was allowed for 5 minutes to dry. A forceps sterilized by flaming was used to press the disc gently on the surface of the agar to promote firm and quick absorption of the antibiotic into the disc. The plates were gently inverted and incubated for 24 hours at 37°C, after which the plates were read the following day to ascertain their zone of inhibition by the antibiotic. Furthermore, the zone of clearance was measured in millilitre (mm) as shown in the result section as described by Cheesbrough [26]. Seven antibiotics were used in this study namely: Tarivid (OFX), Peflacin (PEF), Gentamycin (CN), Augmentin (AU), Ciprofloxacin, (CPX), Septrin (SXT) and Ceporex (CEP)
Statistical analysis
Data were collected using Microsoft Excel and transferred into Statistical Package for Social Science (SPSS) for statistical analysis. Critical consideration was given to statistical analysis SPSS version 21 to analyse the data for mean, standard deviation frequency, percentage, prevalence, ANOVA and correlation. Test of significance was performed at 0.05 alpha levels. Results were presented in tables and charts respectively.
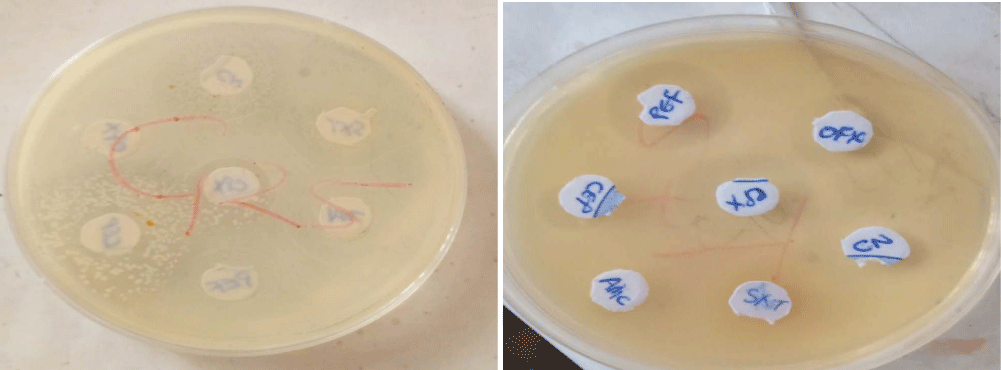
The results obtained from the antibiogram pattern of potential pathogenic bacteria associated with domestic dog faecal matter revealed the presence of Gram-negative rod bacteria, Gram-positive rod bacteria and Gram-positive cocci which showed different reactivity to antibiotics used. See figure 1 below.
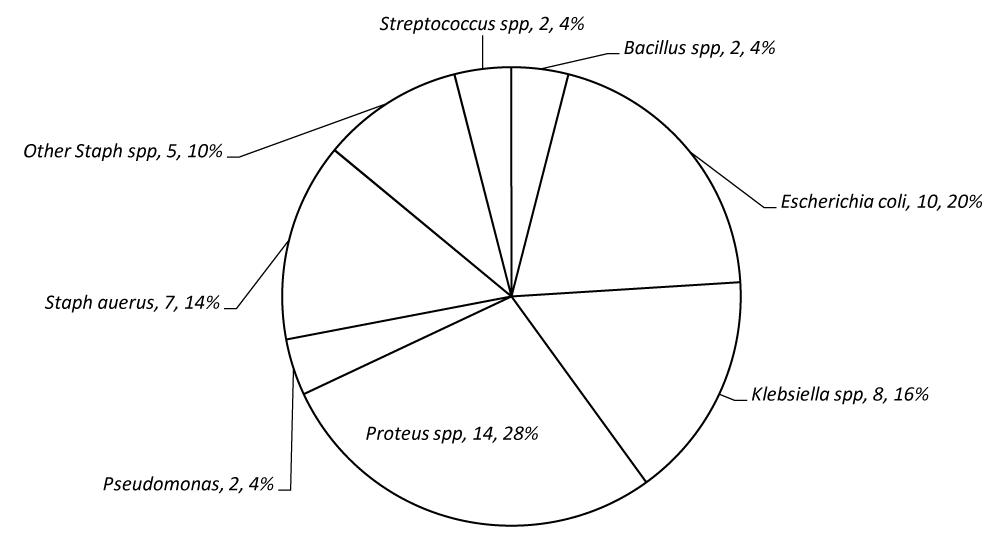
Figure 2 presents Pie Chart showing some Pathogenic Bacteria that are Associated with Domestic Dog Stool at varying frequency namely; Bacillus spp, Escherichia coli, Klebsiella spp. Proteus spp, Pseudomonas spp, Streptococcus spp, Staphylococcus aureus and other Staphylococcus spp. See figure 1 for detail.
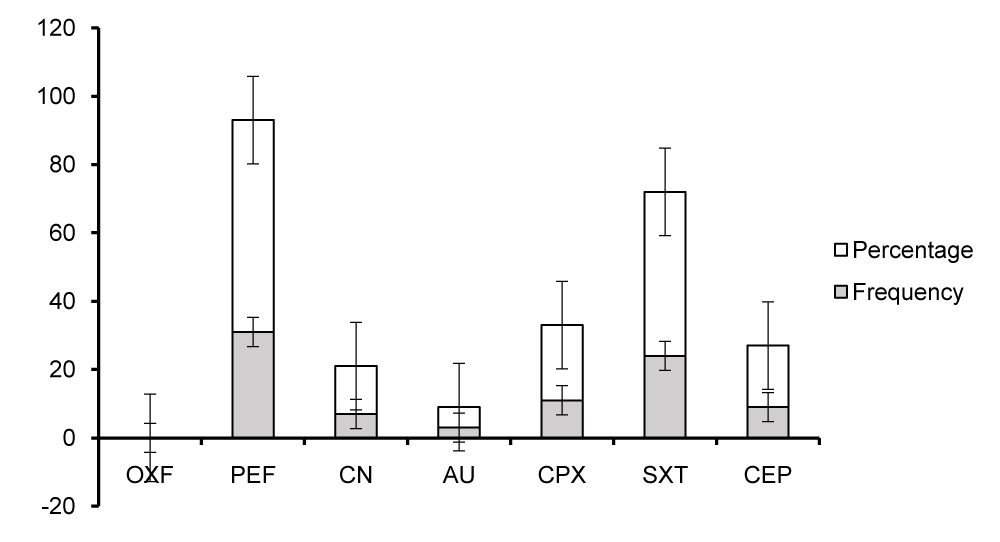
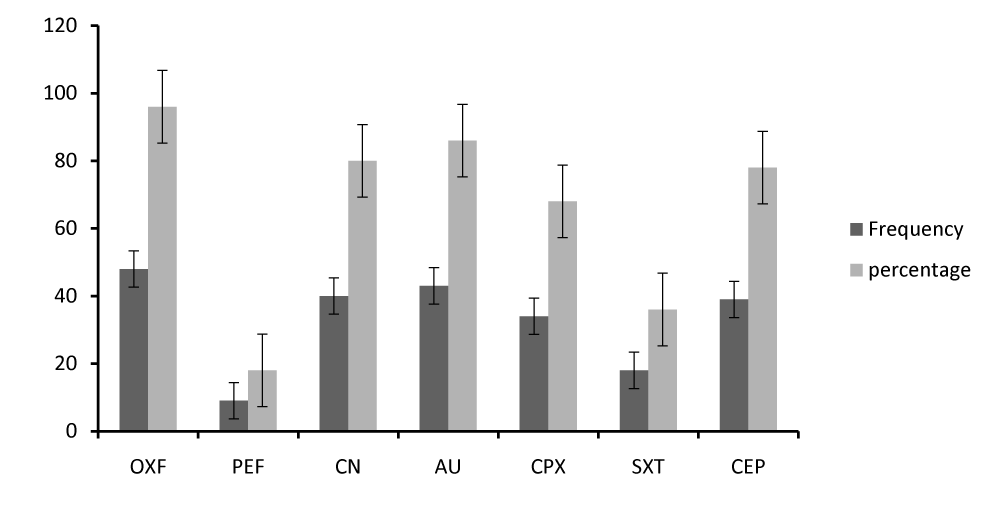
The study further determined the prevalence of antibiotics resistance and susceptibility of the potentially pathogenic bacteria associated with domestic dog stool. Generally, the study showed the frequency distribution of antibiogram of sensitivity profile of different antibiotics used that; 96.0% were sensitive to OXF, 4.0% were moderate to it and 0% were resistant to it. 62.0% were resistant to PEF, 20.0% moderate to it and 18.0% sensitive to it. 80.0% were sensitive to CN, 14.0% resistant and 6% moderate to it. 86.0% were sensitive to AU, 8.0% moderate and 6.0% resistant to it. 68.0% were sensitive to CPX, 22.0% resistant, 10.0% moderate to it. 48.0% were resistant to SXT, 36.0% sensitive and 16.0% moderate to it. 78.0% were sensitive to CEP, 18.0% resistant and 4.0% moderate to it.
Figure 3 shows the prevalence of antibiotic resistance of the different antibiotics used. It indicates that PEF is the most resistant antibiotic (62.0%), while AU is the least resistant antibiotic (6.0%) and OXF has 0% resistant.
Figure 3 presents the prevalence of antibiotic sensitivity of the different antibiotics used. It specifies that OXF is the most sensitive antibiotic (96.0%), followed by AU (86.0%), CN (80.0%), CEP (78.0%), CPX (68.0%), SXT (36.0%), while PEF is the least sensitive (18.0%).
In addition, this study compared the mean difference of the minimum zone of inhibition (antibiogram) of the various potential pathogenic bacteria that are associated with domestic dog stool in Port Harcourt Metropolis.
Table 1 and table 2 present the mean comparison of the antibiotic sensitivity. Table 1 specifically shows the descriptive statistics of mean, standard deviation and number sampled of the minimum zone of inhibition measured while table 2 gives an idea about the ANOVA of antibiotics sensitivity of various isolates based on the minimum zone of inhibition exhibited. The organisms, when tested on PEF, CN, CPX, SXT and CEP, showed significant difference (p < 0.05) in the Mean values of the minimum zone of inhibition (antibiogram) between the isolates. This means that the bacteria isolated in this study possess dissimilar antibiotic profile for the peflocin, gentamycin, ciprofloxacin, septrin and ceporex. However, drugs like Tarivid (OFX) and Augmentin (AU) proved no significant difference (p > 0.05) as obtained from the mean difference of the minimum zone of inhibition (antibiogram) of the various Potential Pathogenic Bacteria associated with Domestic Dog Stool. This connotes that tarivid and augmentin had equal effects on the various organisms.
Table 1: Descriptive Statistics of Antibiotic Sensitivity (Zone of Inhibition). |
||||||||
| Isolates | Tarivid | Peflocin | Gentamycin | Augmentin | Ciprofloxacin | Septrin | Ceporex | |
| Bacillus spp | Mean (N = 2) | 28.00 | 12.00 | 25.00 | 26.00 | 17.00 | 20.00 | 15.00 |
| Std. Deviation | 2.828 | 16.971 | 4.243 | 2.828 | 7.071 | 14.142 | 7.071 | |
| Escherichia coli | Mean (N = 10) | 23.10 | 6.60 | 24.40 | 19.00 | 7.40 | 19.40 | 9.00 |
| Std. Deviation | 3.542 | 6.670 | 2.459 | 11.205 | 11.965 | 8.435 | 12.587 | |
| Klebsiellaspp | Mean (N = 8) | 24.50 | 23.50 | 5.25 | 25.63 | 24.75 | 7.88 | 26.00 |
| Std. Deviation | 3.162 | 9.547 | 11.361 | 3.378 | 3.955 | 9.628 | 3.207 | |
| Proteus spp | Mean(N = 14) | 23.64 | 3.00 | 25.14 | 26.57 | 20.71 | 2.07 | 25.43 |
| Std. Deviation | 2.205 | 4.946 | 2.878 | 3.204 | 11.599 | 5.553 | 5.170 | |
| Pseudomonas | Mean (N = 2) | 24.00 | 17.00 | 17.50 | 22.00 | 13.00 | 24.00 | 17.50 |
| Std. Deviation | 2.828 | 9.899 | 6.364 | 2.828 | 18.385 | 5.657 | 4.950 | |
| Staph aureus | Mean (N = 7) | 23.71 | 3.14 | 24.14 | 24.14 | 21.57 | 16.57 | 23.14 |
| Std. Deviation | 2.138 | 5.398 | 2.734 | 4.413 | 4.614 | 7.892 | 4.140 | |
| Other Staph spp | Mean (N = 5) | 22.60 | 4.80 | 24.20 | 22.80 | 22.60 | 20.00 | 22.60 |
| Std. Deviation | 2.608 | 6.573 | 3.033 | 3.347 | 4.099 | 5.831 | 3.435 | |
| Streptococcus spp | Mean (N = 2) | 25.00 | 6.00 | 26.00 | 25.00 | 24.00 | .00 | 16.00 |
| Std. Deviation | 4.243 | 8.485 | .000 | 4.243 | 5.657 | .000 | 19.799 | |
| Total | Mean (N = 50) | 23.82 | 8.24 | 21.30 | 23.92 | 18.68 | 11.80 | 20.52 |
| Std. Deviation | 2.819 | 9.968 | 8.770 | 6.272 | 10.737 | 10.751 | 9.643 | |
Table 2: Mean Comparison (analysis of variance) of antibiotic sensitivity (zone of inhibition). |
||||||||
| Sum of Squares | df | Mean Square | F | Sig. | Remark | |||
| Tarivid* Isolates | Between Groups | (Combined) | 54.637 | 7 | 7.805 | .979 | .459 | N/S |
| Within Groups | 334.743 | 42 | 7.970 | |||||
| Total | 389.380 | 49 | ||||||
| Peflocin* Isolates | Between Groups | (Combined) | 2707.063 | 7 | 386.723 | 7.512 | .000 | Sig |
| Within Groups | 2162.057 | 42 | 51.478 | |||||
| Total | 4869.120 | 49 | ||||||
| Gentamycin* Isolates | Between Groups | (Combined) | 2562.729 | 7 | 366.104 | 12.752 | .000 | Sig |
| Within Groups | 1205.771 | 42 | 28.709 | |||||
| Total | 3768.500 | 49 | ||||||
| Augmentin* Isolates | Between Groups | (Combined) | 388.719 | 7 | 55.531 | 1.516 | .188 | N/S |
| Within Groups | 1538.961 | 42 | 36.642 | |||||
| Total | 1927.680 | 49 | ||||||
| Ciprofloxacin * Isolates | Between Groups | (Combined) | 1887.209 | 7 | 269.601 | 3.010 | .012 | Sig |
| Within Groups | 3761.671 | 42 | 89.564 | |||||
| Total | 5648.880 | 49 | ||||||
| Septrin* Isolates | Between Groups | (Combined) | 3232.082 | 7 | 461.726 | 7.974 | .000 | Sig |
| Within Groups | 2431.918 | 42 | 57.903 | |||||
| Total | 5664.000 | 49 | ||||||
| Ceporex* Isolates | Between Groups | (Combined) | 2094.494 | 7 | 299.213 | 5.104 | .000 | Sig |
| Within Groups | 2461.986 | 42 | 58.619 | |||||
| Total | 4556.480 | 49 | ||||||
p < 0.05 = Significant (*); p > 0.05 = Not Significant. Df = Degree of Freedom. |
||||||||
In table 3, correlation between Antibiogram and the type/species of Isolates showed almost no evidence of statistically significant correlation except for Ciprofloxacin (CPX) which showed a moderate correlation (0.322, p = 0.02) implying a direct relationship. Nonetheless, no indication of a statistically significant correlation exists between antibiogram and bacterial count. But the study recorded a marked inverse relationship between isolates and bacterial count.
| Table 3: Correlation Analysis between Antibiogram and Types of Isolates withBacterial Count | ||||||||||
| Isolates | Bacterial Count | OFX | PEF | CN | AU | CPX | SXT | CEP | ||
| Types of Isolates | Correlation | 1 | -.405** | -.101 | -.257 | .195 | .110 | .322* | -.051 | .277 |
| p-value | .004 | .487 | .072 | .174 | .447 | .023 | .727 | .052 | ||
| Bacterial Count | Correlation | -.405** | 1 | .049 | .024 | .146 | .073 | -.079 | -.042 | .017 |
| p-value | .004 | .734 | .867 | .311 | .612 | .584 | .773 | .905 | ||
| **.Correlation issignificant at the 0.01 level (2-tailed). *. Correlation is significant atthe 0.05 level (2-tailed). | ||||||||||
Dog faecal matter in urban or rural settings may represent an important source of potential pathogenic microorganisms that would contaminate both dog owners and the community ecosystem at large, thus making the environment unsafe for human habitation. Nonetheless, Kuivusilta & Colleague [27] revealed that infections are transmitted by domestic dogs including Pet dogs with different causative agents in our communities. The observation from this study contradicts the report of a study carried out on dog faeces in the University of Bari, Italy which reported no presence of Salmonella, Yersinia or Campylobacter species isolated from dog faecal matter. However, in another development, Giardia cysts were detected in (1.9%) the faecal samples. Enterococcus species were found to be predominant, E. faecium (61.6%), E. gallinarum (23.3%) and E. casseliflavus (5.5%) respectively [27]. This is in agreement with the present study that the faecal matter of dog pets could harbour some pathogenic bacteria of clinical importance.
Furthermore, Rodrigues and colleagues [28] also reported that dogs and dog faecal matter have been implicated to be carriers of antibiotics resistant bacteria strains [28]. In particular, the presence of Vancomycin-Resistant Enterococci (VRE) in pet animals, including dogs, has been reported [29]. The strains isolated from another study were also found to be resistant to some antibiotics such as clindamycin (86.3%), tetracycline (65.7%), Erythromycin (60.27%) and Ampicillin (47.9%) at varying degree of percentages. High-Level Aminoglycoside Resistance (HLAR) was found in 65.7% of enterococci [30]. Similarly, resistance to three or more antibiotics and six or more antibiotics were observed in this current study likewise the report of Cinquepalmi and colleagues [31] which are in consonance with the present study.
Furthermore, a previous study highlighted the antibiotics sensitivity results pattern that could be used for treating enteropathogens in dog faecal matter originated infection, thus with gentamycin being the most sensitive 95%, followed by azithromycin 50%, enrofloxacin 25% and cefotaxime 20%. This is not in agreement with this study since the most sensitive antibiotic was tarivid 96%, followed by augmentin 86%, gentamycin 80%, ceporex 78%, ciprofloxacin 68%, septrin 36% and peflacine 18% [32]. This difference in the sensitivity pattern may be as a result of different genes found in these pathogens and the presence of plasmid in their cells. It could also be as a result of excess and indiscriminate consumption of antibiotics most times without physician prescription and monitoring, especially in developing communities were citizens combines the treatment of illness with native herbs arbitrarily without recourse of minding the public health implication of such a high-risk practice towards multiplying the burden of antibiotic-resistant strains pathogens in our already weak health facilities and communities in general. Also from the work, peflacine was the most resistant antibiotic of enteropathogens in dogs in the studied area which probably could be linked to drug abuse practice and inconsistencies in playing to the rules guiding the use and application of antibiotic drugs in infectious disease control strategies. Nevertheless, it is probably believed that the occurrence of the antibiotic resistance pathogens may also arise as a result of lack of veterinary clinic and trained personals to treat the dogs hence, most of the dog owners treat their pets by themselves without the experience of health practice skills on how to administer and manage the health of the domestic pet dogs when they are sick.
From the research, it is also known that most of the pets are resistant to antibiotics, which pose a general threat to the community thereby putting the public at risk of getting infected. Therefore, it is important that these domestic pets should be treated and vaccinated properly. Also, pets and humans should not be allowed to eat in the same plate or sleep on the same bed as these served as a major source of transmitting zoonotic infections to man.
Lastly and most interestingly, an examination of the correlation between potential pathogenic bacteria isolated from domestic dog stool with the bacteria count and antibiogram pattern were evaluated. The study utilized the correlation analysis as one of the test statistics to make inferences. The rationale behind this correlation analysis stems from the establishment of a relationship between variables understudy, not causality. Two correlation analysis were obtained in this study and it revealed; the correlation between Antibiogram and Types/species of Isolates (Bacteria) also; the correlation between Antibiogram and Bacterial Count. The study focused on the association between the antibiotic profile in relation to the types/species of Bacteria isolated (qualitatively) and the bacterial count (which is a measure of the load-quantitative). The fact that some bacterial species or types are susceptible to some types of antibiotics and the issue of antibiotic resistance seen in recent time made the researchers look in this direction. Thus, in this study relationship between the bacteria species identified and the amount as revealed by bacterial count were both correlated against antibiogram assuming the null hypothesis of no relationship between the variables. These was a remarkable insight and outcome of the importance of correlation analysis in this present study
Furthermore, some bacteria associated with dog stool isolated in this study are directly related to the antibiogram pattern obtained for ciprofloxacin (0.322) on a moderate level. It means that as the bacteria isolates are increased; the amount/dose of CPX that will bring about therapeutic effect needs to be increased as well. On the other hand, a reduction in the Bacteria associated with dog stool isolated here will not require a high dose of CPX rather a commensurate or reduced level of CPX is required also to achieve potency.
The study revealed the antibiogram pattern of different antibiotics used against the isolated microorganisms. It showed among the different antibiotics, tarivid was most sensitive, while peflacine was the least sensitive. It also revealed that peflacine was the most resistant, while augmentin was the least resistant. This showed that dogs carry potential pathogenic organisms present in stool and this can serve as a source of infection to the pet owners, especially the strains showing resistance to antibiotics. Since domestic dog stool and faecal matters are exposed to the environment, it poses a global public health threat; putting the general public at risk of contracting infectious microorganisms that show resistance to antibiotics.
We sincerely wish to thank all the pet dog owners who permitted us to collect samples from their pet animals throughout the duration of practical bench work. We are also grateful to some veterinary medicine practitioners who assisted us during the sample collection in their various clinics. We are indebted to Prof. S.D Abbey and Prof. (Mrs) G.N Woken and Dr Azuonwu Good luck for their moral support and sense of encouragement. We are thankful to all.
- World Health Organization. Animal Bites. 2018. https://tinyurl.com/y7br4b4w
- Richard L Oehler, Ana P Velez, Michelle Mizrachi, Jorge Lamarche, Sandra Gompf. Bite-related and septic syndromes caused by cats and dogs. Lancet Infect Dis. 2009; 9: 439-447. DOI: 10.1016/S1473-3099(09)70110-0
- Center for Disease Control. Animal-human disease (zoonosis). 2014.
- Marks SL, Rankin SC, Byrne BA, Weese JS. Enteropathogenic bacteria in dogs and cats: Diagnosis, epidemiology and control. J Vet Intern Med. 2011; 125:1195-1208. DOI: 10.1111/j.1939-1676.2011.00821.x
- Claerebout E, Casaert S, Dalemans AC, De Wilde N, Levecke B, Vercruysse J, et al.Giardiaand other intestinal parasites in different dog populations in Northern Belgium. Vet Parasitol. 2009; 161:41-46. DOI: 10.1016/j.vetpar.2008.11.024
- AurélienGrellet, Alexandre Feugier, Sylvie Chastant-Maillard, Bruno Carrez, CorineBoucraut-Baralon, Gregory Casseleux, et al. Validation of a fecal scoring scale in puppies during the weaning period. PrevVetMed. 2012; 106: 315-323. DOI: 10.1016/j.prevetmed.2012.03.012
- Leena K Koivusilta, AnsaOjanlatva. To have or not to have a pet for better health. PLoSOne, 2016; 1: e109. DOI: 10.1371/journal.pone.0000109
- VittoriaCinquepalmi, Rosa Monno, Luciana Fumarola, GianpieroVentrella, Carla Calia, Maria Fiorella Greco. Environmental contamination by dog feces: A public health problem. Int J Environ Res Public Health. 2012; 64:72-84. DOI: 10.3390/ijerph10010072
- Rodrigues J, Poeta P, Martins A, Costa D. The importance of pets as a reservoir of resistant Enterococcus strain with special reference to vancomycin. Journal of Veterinary Medicine Series B. 2002; 49: 278-280. DOI: 10.1046/j.1439-0450.2002.00561.x
- VittoriaCinquepalmi, Rosa Monno, Luciana Fumarola, GianpieroVentrella, Carla Calia, Maria Fiorella Greco, et al. Environmental contamination by dog feces: A public health problem. Int J Environ Res Public Health. 2012;64:72-84. DOI: 10.3390/ijerph10010072
- Peter Damborg, Anne H Sørensen, Luca Guardabassi. Monitoring of antimicrobial resistance in healthy dogs: First report of canine ampicillin-resistant Enterococcus faecium clonal complex 17. Vet Microbiol. 2008; 132: 190-196. DOI: 10.1016/j.vetmic.2008.04.026
- Dukki Han, Tatsuya Unno, Jeonghwan Jang, Kyungtaek Lim, Sun-Nim Lee, GwangPyoKo. The occurrence of virulence traits among high-level aminoglycosides resistant Enterococcus isolates obtained from feces of humans, animals, and birds in South Korea. Int J Food Microbiol. 2011; 144: 387-392. DOI: 10.1016/j.ijfoodmicro.2010.10.024
- Mahony RO, Abbott Y, Leonard FC, Markey BK, Quinn PJ, Pollock PJ. Methicillin-Resistant Staphylococcus Aureus (MRSA) isolated from animals and veterinary personel in Ireland. Vet Microbiol. 2005; 109: 285-296. DOI: 10.1016/j.vetmic.2005.06.003
- Farrin A Manian. Asymptomatic nasal carriage of mupirocin-resistant (MRSA) in a pet dog associated with MRSA infection in household contact. Clin Infect Dis. 2003; 36: e26-8. DOI: 10.1086/344772
- Karen A Moriello. Zoonotic skin diseases of dogs and cats. Animal Health Research Reviews. 2003; 4: 157-168. DOI: 10.1079/AHR200355
- Finola Leonard. Salmonella infection and carriage: The importance of dogs and their owners. VetRec. 2014; 174: 92-93. DOI: 10.1136/vr.g367
- Leonard EK, Pearl DL, Finley RL, Janecko N, Peregrine AS, Reid-Smith RJ, et al. Evaluation of pet‐related management factors and the risk of Salmonella spp. Carriage in pet dogs from volunteer households in Ontario (2005-2006). Zoonoses Public Health. 2011;58: 140-149. DOI: 10.1111/j.1863-2378.2009.01320.x
- Clarence Bagshaw, Allen E Isdell, Dharma S Thiruvaiyaru, Lehr Brisbin I Jr, Susan Sanchez. Molecular detection of canine parvovirus in flies (Diptera) at open and closed canine facilities in the eastern United State. Prev Vet Med. 2014; 114: 276-284. DOI: 10.1016/j.prevetmed.2014.02.005
- Ahmed Garba, Oboegbulem, Junaidu, AbdullahiMagaji, Umoh, Ahmed A, Danbirni S, et al. Rabies virus antigen in the brains of apparently healthy slaughtered dogs in Sokoto and Katsina States, Nigeria. Nigerian Journal of Parasitology. 2010;31:123-125. DOI: 10.4314/njpar.v31i2.69483
- Veronica O Ameh, Asabe A Dzikwi, Jarlath U Umoh. Assessment of knowledge, attitude and practices of dog owners to canine rabies in Wukari Metropolis, Taraba State. Nigeria. Glob J Health Sci. 2014;6:226-240. DOI: 10.5539/gjhs.v6n5p226
- Leslie E Odeh, Jarlath U Umoh, AsabeAdamuDzikwi. Assessment of risk of possible exposure to rabies among processors and consumers of dog meat in Zaria and Kafanchan, Kaduna State. Nigeria. Glob J Health Sci. 2014;61:142-153. DOI: 10.5539/gjhs.v6n1p142
- Umoh JU, Ezeokoli CD, Okoh EAJ. Rabies and rabies-related viruses. Viral diseases of animals in africa. Olufemi Williams A, Masiga WN, editors. OAU/STRC scientific publication. 1988.
- B Hoffmann, Freuling CM, Wakeley PR, Rasmussen TB, Leech S, Fooks AR, et al. Improved safety for molecular diagnosis of classical rabies viruses by use of a TaqMan Real-Time Reverse Transcription- PCR 'Double Check'Strategy. J ClinMicrobiol. 2010; 48:3970-3978. DOI: 10.1128/JCM.00612-10
- Ekeinde A. Slum demolition plan ups tension in Nigeria oil hub.Reuter Africa. 2012.
- Peter Damborg, Anne H Sørensen, Luca Guardabassi. Monitoring of antimicrobial resistance in healthy dogs: First report of canine ampicillin-resistant Enterococcus faecium clonal complex 17. Vet Microbiol.2008; 132: 190-196. DOI: 10.1016/j.vetmic.2008.04.026
- Cheesbrough M.District Laboratory Practice in Tropical Countries. 2nd Ed. Cambridge University Press: The Edinburgh Building. 2006.
- Leena K Koivusilta, AnsaOjanlatva. To have or not to have a pet for better health. PLoS One, 2016; 1: e109. DOI: 10.1371/journal.pone.0000109
- Jorge Rodrigues, PatríciaPoeta, Martins A, Deonisio Costa. The importance of pets as a reservoir of resistant Enterococcus strain with special reference to vancomycin. Journal of Veterinary Medicine Series B. 2002; 49: 278-280. DOI: 10.1046/j.1439-0450.2002.00561.x
- Devriese LA, Leven M, Goossens H, Vandamme P, Pot B, Hommez J, et al. Presence of vancomycin-resistant enterococci in farm pet animal. Antimicrob Agents Chemother. 1996; 40: 2285-2287.PubMed: https://pubmed.ncbi.nlm.nih.gov/8891131/
- VittoriaCinquepalmi, Rosa Monno, Luciana Fumarola, GianpieroVentrella, Carla Calia, Maria Fiorella Greco, et al. Environmental contamination by dog feces: A public health problem. Int J Environ Res Public Health. 2012; 64:72-84. DOI: 10.3390/ijerph10010072
- VittoriaCinquepalmi, Rosa Monno, Luciana Fumarola, GianpieroVentrella, Carla Calia, Maria Fiorella Greco, et al. Environmental contamination by dog feces: A public health problem. Int J Environ Res Public Health. 2012; 64: 72-84. DOI: 10.3390/ijerph10010072
- KokilaPriya A, Balagangatharathilagar M, Chandrasekaran D, Parthiban M, Prathaban S. Prevalence of enteropathogens and their antibiotic sensitivity pattern in puppies with haemorrhagic gastroenteritis. Vet World. 2017; 10: 859-863. DOI: 10.14202/vetworld.2017.859-863
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.