Biology Group 2025 May 28;6(5):532-544. doi: 10.37871/jbres2108.
Differential Proteomic Profile in the Amniotic Fluid of Pregnancies with Trisomy 21
Raquel Rodríguez-López1*, Yaiza Hernández Palomares2, Fátima Gimeno Ferrer3, Irene Ferrer Bolufer1, Carola Guzmán Luján1, Otilia Zomeño Alcalá1, Noelia Cabrera1, María Luz Valero Rustarazo4, José Estardid Colom5, María José Velasco Esteban5, Goitzane Marcaida Benito1, Amparo Secaduras Mora5 and Manue1 M Sánchez del Pino4,6
2Science Faculty, Autonomous University, Barcelona, Spain
3Department of Physiology, Institute of Theoretical Medicine, Medical Faculty, University of Augsburg, Augsburg, Germany
4Proteomics Section, Central Research Support Service (SCSIE), University of Valencia, Spain
5Gynecology Service, Consorcio Hospital General Universitario, Valencia, Spain
6Department of Biochemistry and Molecular Biology, University Institute of Biochemistry and Molecular Biology (BIOTECMED), University of Valencia, Spain
Abstract
Background: The precise analytical-clinical characterization of pregnancies carrying the most frequent aneuploidies has now been surpassed by the technical capacity to obtain their proteomic profile. Amniotic fluid contains the differential proteins related to its specific genetic alteration, representing the molecular etiopathogenesis that generates the associated phenotypes.
Method: The description of the pathognomonic proteomic profiles of amniotic fluid, obtained by Data Independent Acquisition mass spectrometry, of 785 proteins in amniotic fluid from 15 fetuses carrying the most frequent aneuploidies and 15 fetuses with normal combined risk screening.
Results: 119 proteins were clearly overrepresented in the T21 samples and 87 were decreased, 12 encoded by genes located on chromosome 21. The specific proteomic profile of pregnancies carrying Trisomy 21 was based on the combination of the COL6A1, DMBT1, HBB and PRG2 proteins. The proteomic profiles associated with T18 and T13 already suggested specific differential profiles, without reaching statistical significance.
Conclusion: The proteomic analysis of the amniotic fluid defined differentially quantified proteins related to the specific genetic alteration of Trisomy 21, as a molecular etiopathogenesis that generates associated phenotypes based on the overexpression of Extracellular Matrix (ECM) genes and adult hemoglobins, as well as a decrease in coagulation factors, the complement system and immunoglobulin fragments.
Introduction
Amniotic fluid cells have been considered essential for prenatal diagnosis, as they can provide information on fetal genetic abnormalities. Their study is routine, mainly for the detection of aneuploidies that most frequently affect fetuses, those of chromosome pairs 13, 18 and 21, although the exponential development of genetic diagnosis techniques already allows obtaining the complete exome of the fetus when clinically justified [1].
From the beginning of the gestational stage, just twelve days after fertilization, the fetus begins to be covered by Amniotic Fluid (AF) inside the uterus, essential for its normal development. The main function of AF is to protect and surround the embryo, providing the different essential nutrients until the moment of birth [2]. The basis of its biochemical composition is formed by proteins, amino acids, carbohydrates, hormones, lipids and electrolytes [3], being progressively studied in depth and evidencing its much greater complexity; even its concentration varies throughout pregnancy and is mainly related to the functions of the different fetal and amniotic compartments.
As pregnancy progresses, the volume of AF increases proportionally until 37-40 weeks of gestation, when its concentration begins to decrease. It is important to emphasize again that a very important percentage of the proteins that determine the protein profile detected in AF in early stages of gestation are not detected in late stages of pregnancy; at the end of pregnancy, it has been shown that the proteomic profile of amniotic fluid is quite different. The concentration of each of the proteins detected depends on both fetal, placental and/or maternal synthesis and degradation, as well as maternal-fetal exchanges through the placenta. This protein transfer depends on different mechanisms such as active transport, passive diffusion and filtration. Consequently, the concentration of each protein present in AF results from a balance between processes that occur simultaneously and are modified over time as gestation progresses [3].
Changes in the composition of amniotic fluid in pregnancies with clinical diagnoses of chorioamnionitis, preeclampsia, gestational diabetes, intrauterine growth retardation and other pregnancy pathologies related to risk factors due to the mother's lifestyle, including physiological changes between the fetus and the mother, have been widely described and accepted [4]. However, the study of biochemical and/or proteomic changes in amniotic fluid due to the existence of aneuploidies in the fetus is more precarious; little by little, the literature is accumulating manuscripts that describe the first biochemical markers in amniotic fluid that are pathognomonic, because they are characteristic and differential, of pregnancies with fetuses affected by Down Syndrome due to Trisomy 21 (T21), as well as some biochemical and/or protein markers associated with fetuses carrying Trisomy of chromosome pair 13 (T13) or chromosome 18 (T18) [5]
The main findings related to the detection of differential protein profiles of the proteins detected in the analysis of amniotic fluid samples, between the 10th and 13th week of gestation, when the most frequent aneuploidies are usually diagnosed, compared with the profiles of pregnancies without pathology, were: elevation of the proteins PGBM, AMBP, CA11, CA13, CA15, AMP5, ApoA1 and decrease of the proteins IBP-1 in Trisomy of Chromosome 21. Elevation of ApoA1 and decrease of the proteins Vitamin D Binding Protein, IGHBP-1, TTR in Trisomy of Chromosome 18 [6].
The exhaustive analysis of the protein composition of amniotic fluid is revealing its enormous importance for healthy fetal development. Based on the conclusions that are being drawn, lines of research have emerged that study whether deviations/alterations in the composition of non-pathological pregnancies can also contribute to the appearance of signs and/or symptoms specific to the phenotype associated with the genetic alteration carried by the fetus. Obviously, it is much more common to design lines of work that attempt to relate the differences in the composition of the amniotic fluid of a pregnancy carrying a fetus with aneuploidy, with the specific genetic alteration that this entails. In any case, it is clear that any deviation in the biochemical and/or proteomic composition of the amniotic fluid samples from an ongoing pregnancy is a very plausible sign of the existence of complications/alterations in the pregnancy and/or in the fetus.
The application of proteomic methodologies has long been considered a highly useful strategy for discovering biomarkers in samples such as AF in a sensitive and efficient manner. This work addresses the study of the fetal conditions characteristic of pregnancies in which one of the most frequent fetal aneuploidies has occurred. They have been defined by comparing them with the proteomic characteristics of normal pregnancies, rigorously matched according to the age of the pregnant woman, the gestational age and the fetal sex. In all of them, the existence of other pregnancy pathologies that could modify the proteomic profile characteristic of each type of pregnancy carrying the most frequent chromosomal trisomies was ruled out. We aim to correlate the genotype of these aneuploidies with a differential expression of the protein profile defined by Data Independent Acquisition mass spectrometry (DIA) [5,6].
Methods
Patients
A case-control study was designed that included the cases of 15 pregnant patients whose combined risk assessment screening resulted in pathological values of T21 and/or T13- T18 <1/1500. In all of them, amniocentesis was indicated for prenatal study of the most frequent aneuploidies: Trisomy 21 (T21), Trisomy 13 (T13), Trisomy 18 (T18) and sex chromosomes recurrent alterations. Of the 15 patients included, 11 of them carried fetuses with chromosomal abnormality of T21 (Down syndrome), 3 of them with T13 (Patau syndrome) and 1 of them with T18 (Edwards syndrome). From the results obtained from the genetic diagnostic tests using the standardized QF-PCR technique, it was concluded that in all patients with chromosomal abnormalities, the error occurred during the first meiotic division. These data are in agreement with the reviewed literature, where most errors occur in the first division [7,8]. As a control series to compare with the pathological cases, we selected another 15 cases of pregnant women whose combined risk calculation screening resulted in parameters in the unquestionable “no risk” range. The cases were matched with the controls according to the age of the pregnant woman and the sex of the fetus. The ages of the pregnant women included in the series ranged between 27 and 42 years. The sex distribution in the fetuses was 7 male and 9 female.
Risk assessment by combined screening
Normal variables for the calculation of the combined risk were considered to be maternal serum beta-free human chorionic gonadotropin (β-hCG), pregnancy-associated plasma protein A (PAPP- A), nuchal translucency (NT), and the ethnicity of the pregnant woman, together with the personal medical history of the pregnant woman. Data such as obesity, biochemical alterations (hypothyroidism, diabetes) and/or pathologies that appeared during pregnancy (low/moderate arterial hypertension, preeclampsia) were taken into account. In assessing the risk of pregnancy, the risk attributed to the age of the pregnant woman is multiplied by the likelihood ratios assigned to the deviation of NT, free β-hCG and PAPP-A measurements from the expected medians, respectively. Regarding the determination of free β-hCG and PAPP-A, their measured serum concentrations were adjusted according to certain maternal and pregnancy characteristics. Basically, each measured level is first converted into a multiple of the expected normal median (MoM) specific to a pregnancy of the same gestational age, maternal weight, ethnicity, smoking status, method of conception, and parity, as well as the autoanalyzer and reagents used for laboratory testing [9].
Early detection of the most frequent chromosomal aneuploidies
In each case, fetal DNA was extracted from 12 to 15 milliliters of amniotic fluid sample, followed by QF-PCR analysis of sequence fragments specific to the chromosomes of interest: In case of the sexual chromosomes, the SRY gene of the Y chromosome, located in the Yp11.31 band, X- and Y- chromosome-specific fragments of the AMEL gene (region Xp22.22/Yp11.2), and the pseudoautosomal region ZFXY (region Yp11.31/Xp22.11). In addition, hypervariable genetic markers between individuals and specific to the X chromosome are amplified: DXS1187, DXS2390 and XHPRT, X and Y chromosomes, chromosome 13 (D13S742, D13S634, D13S628, D13S305, D13S1492), chromosome 18 (D18S978, D18S535, D18S286, GATA178F11) and chromosome 21 (D21S1435, D21S11, D21S1411, D21S14444, D21S1442, D21S1437).
Mass spectrometry analysis
The supernatant was preserved from each sample of amniotic fluid extracted for genetic study. Proteins were precipitated in 60 μl of the sample by adding 40 μl of pure ethanol and incubating overnight at 5°C. After centrifugation at 21,000 g and a temperature of 5°C, the pellet was obtained; 40 μl of 50% ethanol was added again at -20 °C and centrifuged to discard the supernatant and air dry the pellet; the pellets were resuspended with 30 μl of RIPA buffer and the protein concentration obtained was measured according to the Qubit Broad Range method. For a concentration of 10 μg of protein, it was diluted to 30 μl with 50 mM ammonium bicarbonate and reduced by adding 10 μl of 2 mM DTT and incubating for 20 minutes at 60 °C. Free thiols were alkylated by adding 10 μl of 5.5 mM iodoacetamide and incubating for 30 min at room temperature in the dark. Protein digestion was carried out by adding 5 μl of trypsin (Promega Sequencing grade at 100 ng/μl) and incubating overnight at 37 °C. The reaction was stopped by adding 5 μl of 10% TFA.
For LC-MS/MS analysis, 1.5 μl aliquots of the digested mixtures were prepared with 20 μl of 0.1% formic acid and loaded onto Evotips (Evosep) according to the manufacturer's instructions. Chromatographic separation of peptides was achieved using the 60 SPD method on an Evosep system coupled to a timsTOF flex mass spectrometer (Bruker). Peptides were ionized in a captive spray with 1600 V at 180 °C and the mass spectrometer was set to the parameters: TIMS, custom mode; 1/K0, 0.6-1.6 V.s/cm2; 100 ms ramp time; duty cycle set to 100%; 9.42 Hz ramp rate; ms averaging set to 1; auto calibration set to off. MS, 100-1700 m/z scan range; positive ion polarity; scan mode set to diaPASEF.
The PASER system (Bruker) was used to submit data for quantification to DIA-NN v 1.8.1 [10]. An in silico predicted spectral library was constructed from a SwissProt Human database against which diaPASEF raw files were analyzed. DIA-NN settings were as follows: cysteine carbamidomethylation as a fixed modification and methionine oxidation as variable modification, with a maximum number of variable modifications set to 1. Trypsin missed-cleavages were set to 1. The peptide length, precursor m/z and charge were set to 7-30, 300-1800, and 1-4, respectively. Analysis was performed in two passes considering all samples as part of the same experiment. The remaining parameters were the default settings. Single genes and protein groups with an FDR ≤ 1% were quantified and normalized using the MaxLFQ algorithm [11].
Data evaluation and statistical analysis
Data quality was assessed by checking the number of proteins quantified per sample, and was considered inadequate when the number of proteins deviated more than 1.5 standard deviations from the average of proteins identified in the overall sample. Quantified proteins were filtered based on their detection frequency and matching peptides. Only proteins with 2 or more peptides that were present in at least 50% of the samples from the control group or the group affected by Trisomy 21 or in all samples from fetuses affected by Trisomy 18 were considered for further analysis.
For statistical analysis, quantitative protein values were transformed into log2. A significance test was performed (Student's T test, bilateral without assuming equal variances). A p value was obtained for each protein if it was detected in at least 2 samples from each of the groups being compared. To select differential proteins, those with a p-value less than 5% were considered. When comparing samples from the Trisomy 21 group with control samples, proteins absent or only present in one sample from one of the groups were considered differential if they were detected in more than 50% of the samples from the other group.
To select those proteins that best defined the control and Trisomy 21 samples, we fitted the data to a logistic regression model. LASSO regularization and five-fold cross- validation were applied using Orange software [12].
Functional analysis
The obtained protein dataset was assigned to the STRING (V12) database using Cytoscape StringApp12 within the Cytoscape software [13]. Differential proteins were selected to create a functional subnetwork and a topological Markov clustering algorithm was used to identify highly connected proteins. Functional enrichment of the clusters was analyzed in Cytoscape StringApp using the entire protein dataset as a basis.
Results
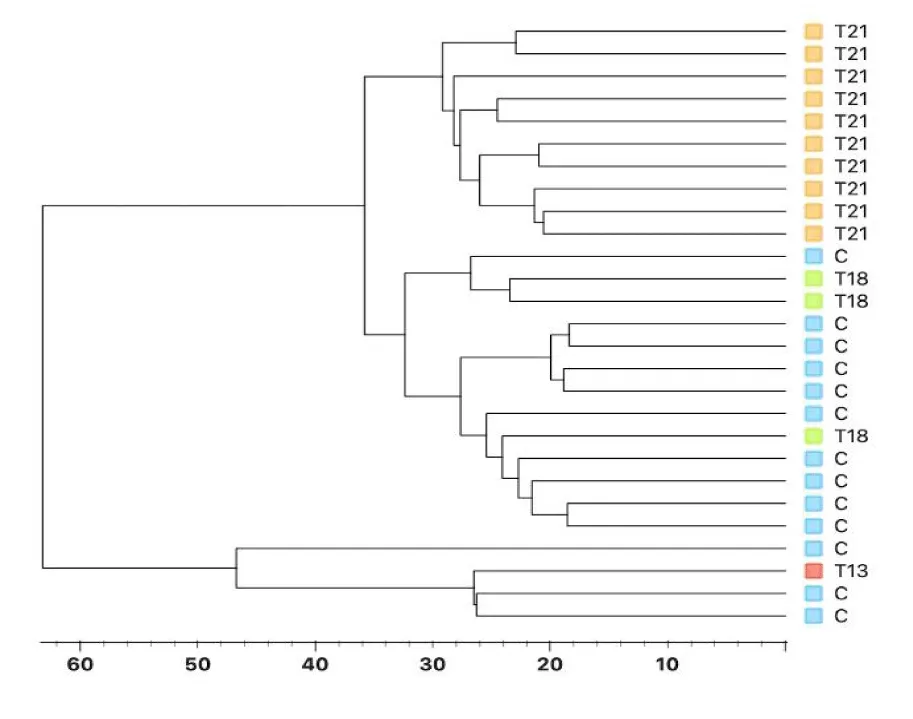
A total of 1189 proteins were identified in the entire series, including cases and controls. In 785 proteins, 2 or more peptides were detected; their existence was confirmed in more than 50% of the samples from the control group or from the group of pregnancies diagnosed with T21, or in all samples from T18 fetuses. This subset of 785 proteins was used for further analysis. Unsupervised clustering of samples using hierarchical clustering, PCA and t-SNE clearly separates samples from the group of pregnancies without pathology (considered control) from samples from pregnancies affected with T21 (Figure 1). Although the number of samples from pregnancies with T18 was very limited, the clustering analysis places these samples at the interface between the regions of the control group and the group with T21, indicating clear differences in their protein profile compared to the other two groups of samples. DIA profile of T21 group versus control group
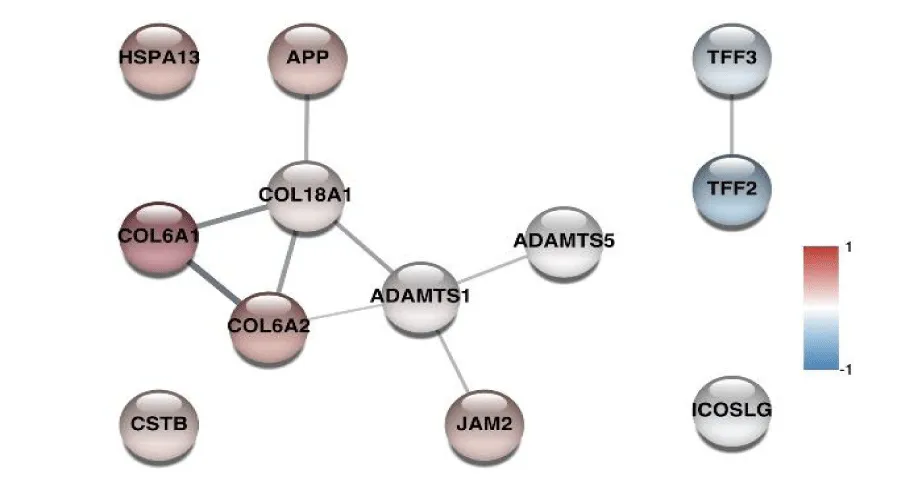
Statistical analysis of the results obtained from the group of fetuses affected by T21 compared to the group of control pregnancies revealed the presence of 206 differentially abundant proteins (DAP), 119 of which were clearly overrepresented in the T21 samples and 87 were decreased (Table 1 of the Supplementary Material). 12 of the identified proteins were encoded by genes located on chromosome 21 (ADAMTS1, ADAMTS5, APP, COL18A1, COL6A1, COL6A2, CSTB, HSPA13, ICOSLG, JAM2, TFF2, TFF3) and 9 of them were found to be higher in the group of samples from pregnancies with T21 (Figure 2).
We used a logistic regression model to fit the data and applied a LASSO regularization from lowest to highest stringency, for feature selection, which was able to classify the samples correctly and revealed a set of 16 proteins under lower stringency conditions (CHI3L1, CLCA1, COL6A1, DCN, DMBT1, HBB, HBD, HBG1, HBG2, IGLV1-47, KRT16, KRT2, NOTUM, PAEP, PLOD2, PRG2) that were reduced to 4 proteins under higher stringency conditions (COL6A1, DMBT1, HBB, PRG2).
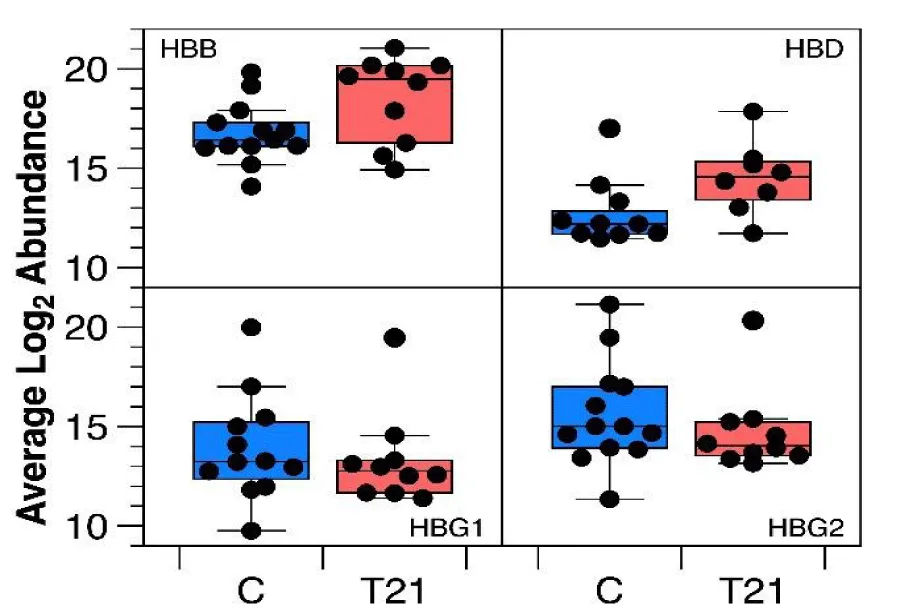
Among this group of proteins, those that make up the beta chains of hemoglobin were highly represented, so a more detailed analysis of the data was carried out and a significant increase in the adult beta chains HBB and HBD could be distinguished in the group of samples from pregnancies with T21, and a decrease in the fetal beta chains HBG1 and HBG2, although they did not reach statistical significance (Figure 3).
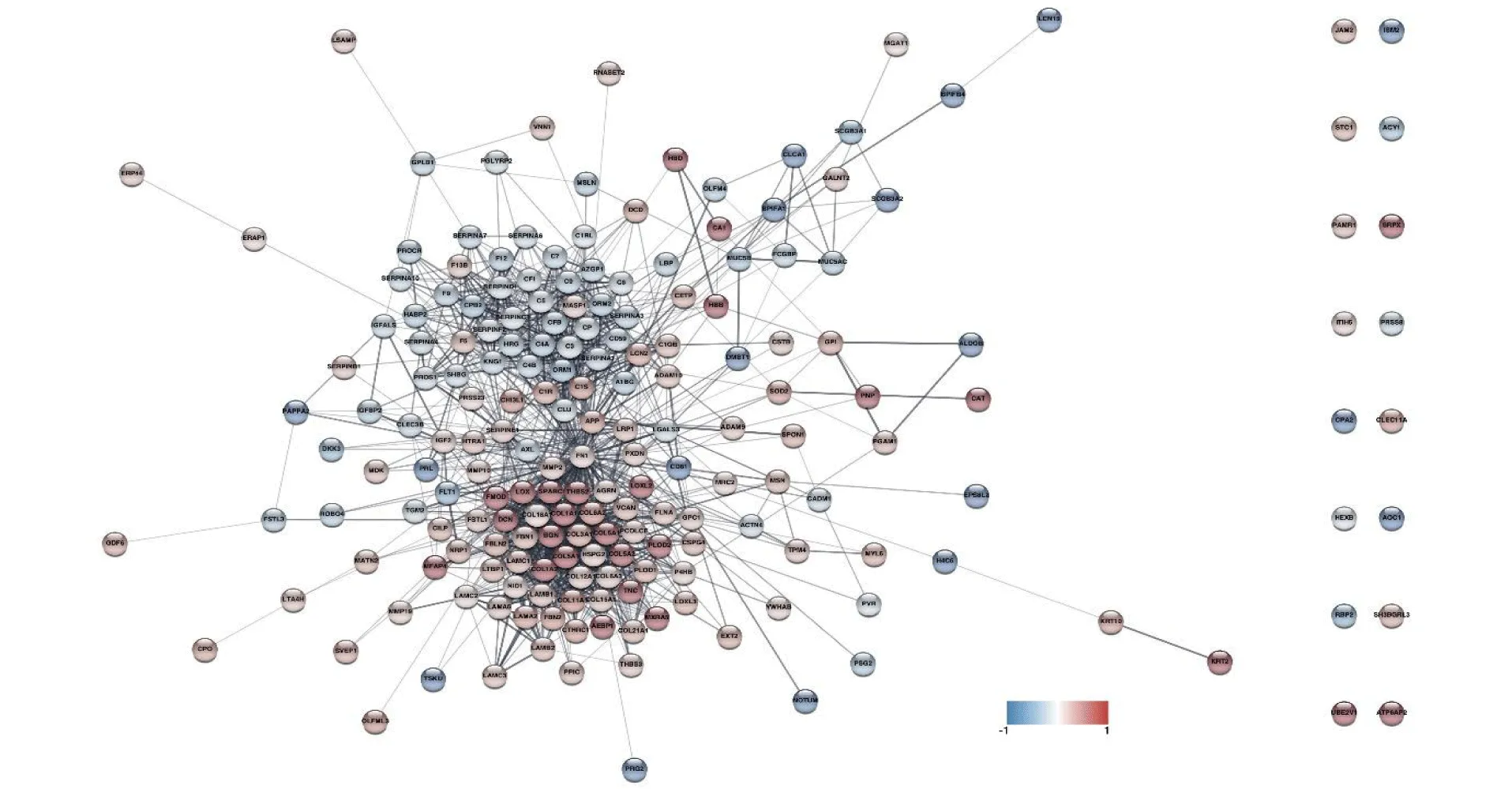
From the 785 identified proteins that met the established criteria, 742 were mapped to the STRING database for functional analysis. The rest were immunoglobulins; analyzed together, they showed a significant decrease in the group of samples belonging to T21 pregnancies. From the mapped proteins, a network of 206 differentially abundant proteins (DAPs) emerged, from which a subnetwork containing 198 DAPs was generated, with which we planned to perform the functional analysis (Figure 4). The results showed a large network of highly connected proteins that contained more than 90% of the DAPs, indicating a high degree of functional correlation between them.
Fibronectin was the central protein which was found connecting both sets of proteins, which were in turn highly connected to each other. The most abundant group of proteins in the group of samples from T21 pregnancies contained the most basic protein regulators that are part of the extracellular matrix (ECM) and related proteins, including 13 types of collagen proteins. The other group of proteins was mainly composed of downregulated proteins in T21 that were enriched in components of the coagulation and complement cascades.
A first approximation of the regulation of the main genes that build the extracellular matrix revealed that their expression is fundamentally modulated by the transcription factor RUNX1, located precisely on chromosome 21. We mapped the extracellular matrix genes that were recognized as regulated by the RUNX1 transcription factor in our dataset, and identified 26 of its encoded proteins. Samples from T21 pregnancies showed a higher abundance of 22 of these 26 proteins (Figure 5).
The advance in the knowledge of collagenopathies/connective tissue disorders-related proteins has revealed their complexity and association with the existence of biologically active fragments called matricryptins and matrikines [14]. We mapped the identified collagen peptides to their corresponding protein sequence (Figure 6), achieving an alignment of some peptides within the sequences of canstatin (COL4A2), restin (COL15A1) and endostatin (COL18A1). The peptides of collagens COL4A2 and COL15A1, in addition to mapping within their corresponding active fragments, also mapped to non-helical domains of other proteins. However, all COL18A1 peptides mapped exclusively within the Endostatin sequence covering approximately 60% of its sequence.
Most of the complement proteins were decreased in the group of samples from T21 pregnancies, except for the classical pathway activators: C1R, C1S and C1q which were clearly elevated; these three proteins showed a high correlation between them. The other group of proteins mostly decreased in T21 were enriched in components of the coagulation cascades (Supplementary Table 1).
Discussion
Proteomic profiles of AF fluid in pregnancies carrying trisomies manifest differential expression set of protein that may affect the development of the fetus during the gestation. In this work is has been shown the different pattern of proteins in T21, that should have a pathophysiological effect associate to newborns with Down Syndrome. The analysis manifested significant changes in proteins which genes were located in chromosome 21, as expected due to the Trisomy, but also of genes like collagen-related and complement associated genes.
The characterization of the proteomic profile of AF generated by fetuses and their appendages when they are carriers of the most frequent aneuploidies in humans, Trisomy of chromosome pair 21, is the basis for understanding the pathophysiology of the phenotype developed by individuals born with associated Down Syndrome or T21 [15].
High-risk screening through noninvasive genetic testing using free circulating DNA analysis of the fetus in a peripheral blood sample of the pregnant woman is undoubtedly the definitive trend that seems to be extended to all pregnancies with any clinical risk, and it is even proposed to establish it as a screening for the general population. Proteomic description from the earliest stages of the affected individual therefore offers data to delve deeper into the true nature of the genetic alteration and provides the option of proposing highly specific clinical management strategies. It may also manifest a different development of fetuses, since different composition of AF may also influence in the fetus itself.
The first subset of 785 proteins identified in the complete series with robust confidence parameters already indicates the differentiating and specific characteristics of the different proteomic profiles shown by AF of each of the three most frequent autosomal aneuploidies (Figure 1); we focused on the in-depth study of the profile of T21, because the sample size of the series of pregnancies with T18 or T13 was very limited and because of the prevalence of the T21 compared the other trisomies.
Most of the 43 unmapped proteins in the STRING database turned out to be immunoglobulins that collectively showed a clear decrease in T21-affected pregnancies. The immunological dysfunction that occurs in Down Syndrome has been widely described, and the correlation with the decrease of the peptides described here could explain and help to prevent both the increased susceptibility to infections and the autoimmune manifestations that affect these patients [16].
Among the identified proteins, only their genes ADAMTS1, ADAMTS5, APP, COL18A1, COL6A1, COL6A2, CSTB, HSPA13, ICOSLG, JAM2, TFF2, TFF3 are located on chromosome 21, but the greater abundance of 9 of them was clearly confirmed in amniotic fluid samples from pregnancies with fetuses carrying three copies of chromosome 21.
Statistical analysis defined the characteristic proteomic profile of pregnancies affected by Trisomy 21, based on the highly significant changes in the 16 proteins CHI3L1, CLCA1, COL6A1, DCN, DMBT1, HBB, HBD, HBG1, HBG2, IGLV1-47, KRT16, KRT2, NOTUM, PAEP, PLOD2, PRG2, being the combination of COL6A1, DMBT1, HBB and PRG2 the most representative. The greater abundance of COL6A1 protein in amniotic fluid of pregnancies with fetuses carrying T21 compared to control samples had already been published [17,18]. The DMBT1 protein showed a clear decrease, as did that encoded by the PRG2 gene, one of the most expressed genes during human pregnancy. The complete form acts as a proteinase inhibitor, reducing the activity of PAPP-A (UniProt information: P13727) whose low levels predict Down syndrome and a poor pregnancy prognosis. These data fit with our results, where it is observed that in T21 fluids, there is a decrease in the protein encoded by the PRG2 gene [19].
The new and robust finding of the significant increase in adult beta chains HBB and HBD in the T21 pregnancy sample group suggests that the change from fetal to adult hemoglobin in T21 was at a more advanced stage than in the control samples; correlation between this dataset and comorbidity of T21 individuals would be essential for suitable clinical management.
A thorough analysis of the proteomic profile of 742 differential proteins of AF from pregnancies with T21 using the database STRING revealed a clear increase in proteins that form the extracellular matrix (ECM) and a clear decrease in proteins involved in coagulation and the complement system, therefore a putative malfunctioning of the immune system. Fibronectin remained as the fundamental protein connecting both groups of proteins, which were highly connected to each other (Figure 4). The overexpression of ECM genes in fetal tissues affected with T21 had already been published and has been previously reported in cardiac fetuses with T21 [20], being our study another confirmation of this observation. Our study identified 26 ECM proteins regulated by RUNX1 [21] in the both T21 and control AF samples, and [22] of them were more abundant in the samples from the T21 group (Figure 5).
The transcription factor RUNX1 may explain many of the alterations in the protein profiles observed in amniotic fluid. RUNX1, whose gene is precisely located on chromosome 21, is an important regulator of gene expression acting at several levels. On the one hand, RUNX1 directly regulates the expression of certain genes, many of them related to the ECM, which explains the increased abundance of ECM proteins described above. In addition, collagen-derived matricryptins and matrikines can modulate morphogenesis and tissue remodeling [23]. Indeed, our data show an increase in endostatin, derived from collagen COL18A1, which has a strong anti-angiogenic activity (below).
On the other hand, RUNX1 is also an essential player in a more global process that coordinates the embryonic hematopoietic program. Overexpression of RUNX1 has been associated with an increased predisposition of individuals with Down syndrome to develop acute megakaryoblastic leukaemia (AMKL). The initiation of the hematopoietic gene expression program requires chromatin remodeling in the region where the transcription factor Pu.1 and its target Csf1r are located (Hoogenkamp M, Lichtinger M, Krysinska H, Lancrin C, Clarke D, Williamson A, Mazzarella L, Ingram R, Jorgensen H, Fisher A, Tenen DG, Kouskoff V, Lacaud G, Bonifer C. Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood. 2009;114:299-309). It is the process of chromatin opening where RUNX1 plays a critical role. The opening of this chromatin region is the prerequisite for both the developmental process of the different hematopoietic cell lines as well as the hemoglobin switch [22]. Our data show an increase in adult hemoglobin and immune system alterations, including a global decrease in immunoglobulin levels. All these effects are compatible with an overexpression of RUNX1.
The attempt to identify biologically active fragments of Canstatin (COL4A2), Restin (COL15A1) and/or Endostatin (COL18A1) resulted in the successful mapping of peptides to the Endostatin sequence. An elevated plasma concentration of Endostatin had previously been reported in patients affected by Down Syndrome [23].
Clinical Genome Resource Web (ClinGen) has not published curations for COL4A2 (HGNC:2203), COL15A1 (HGNC:2192) yet, but Collagen type XVIII alpha 1 chain (COL18A1) (HGNC:2195) was lastly curated in 2024. Phenotypic variability has been reported in patients carrying pathogenic variants in COL18A1 gene with ocular defects, brain malformations, developmental delay and seizures when the mechanism of pathogenicity is biallelic inherited with loss-of-function variants. The positively co-expression of COL18A1 with other genes implicated in cortical malformation syndromes, and detection of COL18A1 mRNA in pyramidal neurons of all layers of the cortical plate have been corroborated [24]. However, the GTEx database assigns the same main transcript of the COL18A1 protein the lowest expression in the brain, with the aortic and coronary arterial tissues being where the protein expression is highest. It could make sense that the increased abundance of COL18A1 verified in the proteomic profile of T21 fetuses may have negative consequences related to the neurological development, and therefore, phenotype that T21 patients present.
Carbonic anhydrases (CAs) are a large family of zinc metalloenzymes that catalyze the reversible hydration of carbon dioxide. They participate in a variety of biological processes and show a wide diversity in tissue distribution, being most abundantly expressed in the brain with a possible general role in the central nervous system. Previous studies had also shown that CAII levels are increased in the brains of infants and young children with Down Syndrome. Although in our research we have not observed CAII but CAI, the expression pattern is the same [25].
After the most common neurodevelopmental disorder suffered by individuals with Down Syndrome, the most frequently associated abnormalities are cardiovascular defects, gastrointestinal musculoskeletal defects, ocular defects, and respiratory defects [26]; after these, abnormalities such as umbilical hernias or anal atresia are relatively frequent, mostly associated with skeletal abnormalities such as joint laxity, abnormalities of the cranial structures with brachycephaly, with short neck and fingers as well, abnormalities with the cervical and vertebral column that cause a very characteristic short stature [27]. Among the musculoskeletal irregularities, hypotonia with clubfoot is very frequent. The skin of these patients can also be significantly affected [28]. It would be extremely interesting to be able to investigate in depth the possible relationship between the elevation of the group of factors that form the ECM and collagen, indicated by the highly differential proteomic profile from such early prenatal stages of the individual development and phenotype.
In recent years, the enormous importance of the correct functioning of the complement system in neurodevelopment has been accepted. The correlation of the decrease in coagulation factors and the complement system in patients with T21 is even less known, although varied hematological abnormalities [29] and immune dysregulation [30] in Down Syndrome are widely reported. Regarding the fact that we detected that most of the complement proteins were decreased in T21 pregnancies, except for the activators of the classical pathway: C1R, C1S and C1q that were clearly elevated, it has been published that adult patients with T21 show most of the complement proteins elevated [31]. However, other studies have corroborated the depletion of most of the complement proteins among individuals affected by Down Syndrome [32-34].
It has been reported that in patients with Down Syndrome, differential hematopoiesis has been observed, which could be associated with a decrease in immunoglobulins and a possible increase in hemoglobin, specifically related to the effect of the RUNX1 protein, contributing potentially to an increased risk factor for the development of certain types of leukemia [35].
Although Down Syndrome is the most frequent neurodevelopmental disorder in humans, despite the large amount of data that the literature about its signs and symptoms, its phenotypic description is still imprecise, as well as modestly correlated with the possible triplosensitivity of all the genes found on chromosome 21. The information described in the proteomic profile, identifiable from the first weeks of development of individuals, should be able to establish good designs for lines of research to go deeper in this direction. The goal of using the data obtained in our research is its application to the field of individual therapy, including in terms of primary prevention of expected comorbidities and better management during the life of the patients by the healthcare institutions.
The results of the proteomic profiles of the group of samples belonging to pregnancies carrying Trisomy of chromosome pair 18 already suggested a specific combination of differential proteins. The sample belonging Trisomy 13 already stood out above the rest; all of this points to the effectiveness of proteomic analysis to propose lines of study of the true etiopathogenic bases of the associated phenotypes. The T21 cohort was well represented, and due to the clear differences in their proteomic profiles, the data obtained achieved significant statistical power with the number of samples included. We were able to observe these clear differences in the analysis of the first cases, so we decided to include the limited data obtained from the few cases we have of pregnancies with T13 and T18 aneuploidies. We hope to obtain more information about the proteomic profiles in larger series before we can begin to draw conclusions.
Acknowledgement
Our thanks to Professor Alicia Alicia Roque Córdova (https://orcid.org/ 0000-0002-6206-6481), Department of Biochemistry and Molecular Biology, Faculty of Sciences, Autonomous University of Barcelona, for co-directing Yaiza Hernandez Palomares' Final Degree Project and which has marked the beginning of our collaboration.
Funding
The line of research has been funded by the Research Foundation of the General University Hospital Consortium of Valencia, within the framework of the 2017 Annual Call for the López Trigo Awards (RRL, IFB, GMB, ASM), PROMETEO/2022/062 from Generalitat Valenciana and PID2020-119111GB-I00 from the Spanish Ministry of Science and Innovation (MSP).
The study proposes a retrospective analysis of a series of patients who requested and consented to clinical care for prenatal clinical and genetic diagnosis. Since the proposed line of research was approved by the Research Foundation of the General University Hospital Consortium of Valencia, within the framework of the 2017 Annual Call for the López Trigo Awards and to promote the research activity of professionals working at the Center, the project was approved after collecting all the required documentation. It was accepted after checking and fulfilling all the requirements demanded by the regulations and legislation in force according to the members of the Ethics and Research Committee of the General University Hospital Consortium of Valencia.
Supplementary Material
Table 1 Supplementary Material: Statistical analysis of the results obtained from the group of fetuses affected by T21 compared to the group of pregnancies without pathology revealed the presence of 206 differentially abundant proteins, 119 of which were clearly overrepresented in the T21 samples and 87 were decreased.
References
- Fu F, Li R, Yu Q, Wang D, Deng Q, Li L, Lei T, Chen G, Nie Z, Yang X, Han J, Pan M, Zhen L, Zhang Y, Jing X, Li F, Li F, Zhang L, Yi C, Li Y, Lu Y, Zhou H, Cheng K, Li J, Xiang L, Zhang J, Tang S, Fang P, Li D, Liao C. Application of exome sequencing for prenatal diagnosis of fetal structural anomalies: clinical experience and lessons learned from a cohort of 1618 fetuses. Genome Med. 2022 Oct 28;14(1):123. doi: 10.1186/s13073-022-01130-x. PMID: 36307859; PMCID: PMC9615232.
- Park HJ, Cho HY, Cha DH. The Amniotic Fluid Cell-Free Transcriptome Provides Novel Information about Fetal Development and Placental Cellular Dynamics. Int J Mol Sci. 2021 Mar 5;22(5):2612. doi: 10.3390/ijms22052612. PMID: 33807645; PMCID: PMC7961801.
- Fitzsimmons ED, Bajaj T. Embryology, Amniotic Fluid. 2022 Jul 19. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing, Bookshelf ID: NBK541089; 2023.
- Vasani A, Kumar MS. Advances in the proteomics of amniotic fluid to detect biomarkers for chromosomal abnormalities and fetomaternal complications during pregnancy. Expert Rev Proteomics. 2019 Apr;16(4):277-286. doi: 10.1080/14789450.2019.1578213. Epub 2019 Feb 13. PMID: 30722712.
- Kolialexi A, Tounta G, Mavrou A, Tsangaris GT. Proteomic analysis of amniotic fluid for the diagnosis of fetal aneuploidies. Expert Rev Proteomics. 2011 Apr;8(2):175-85. doi: 10.1586/epr.10.112. PMID: 21501011.
- Cho CK, Diamandis EP. Application of proteomics to prenatal screening and diagnosis for aneuploidies. Clin Chem Lab Med. 2011 Jan;49(1):33-41. doi: 10.1515/CCLM.2011.002. Epub 2010 Oct 20. PMID: 20961197.
- Dagna Bricarelli F, Pierluigi M, Grasso M, Strigini P, Perroni L. Origin of extra chromosome 21 in 343 families: cytogenetic and molecular approaches. Am J Med Genet Suppl. 1990;7:129-32. doi: 10.1002/ajmg.1320370726. PMID: 1981472.
- del Mazo J, Pérez Castillo A, Abrisqueta JA. Trisomy 21: origin of non-disjunction. Hum Genet. 1982;62(4):316-20. doi: 10.1007/BF00304546. PMID: 6219937.
- Kagan KO, Wright D, Baker A, Sahota D, Nicolaides KH. Screening for trisomy 21 by maternal age, fetal nuchal translucency thickness, free beta-human chorionic gonadotropin and pregnancy-associated plasma protein-A. Ultrasound Obstet Gynecol. 2008 Jun;31(6):618-24. doi: 10.1002/uog.5331. PMID: 18461550.
- Demichev V, Messner CB, Vernardis SI, Lilley KS, Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat Methods. 2020 Jan;17(1):41-44. doi: 10.1038/s41592-019-0638-x. Epub 2019 Nov 25. PMID: 31768060; PMCID: PMC6949130.
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014 Sep;13(9):2513-26. doi: 10.1074/mcp.M113.031591. Epub 2014 Jun 17. PMID: 24942700; PMCID: PMC4159666.
- Demšar J, Curk T, Erjavec A, Gorup Č, Hočevar T, Milutinovič M, et al. Orange: Data mining toolbox in Python Journal of Machine Learning Research. 2013;14,2349–2353.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003 Nov;13(11):2498-504. doi: 10.1101/gr.1239303. PMID: 14597658; PMCID: PMC403769.
- Ricard-Blum S, Salza R. Matricryptins and matrikines: biologically active fragments of the extracellular matrix. Exp Dermatol. 2014 Jul;23(7):457-63. doi: 10.1111/exd.12435. PMID: 24815015.
- Human Phenotype Ontology Website. 2024.
- Bordon Y. Immune dysregulation in Down syndrome. Nat Rev Immunol. 2023 Apr;23(4):201. doi: 10.1038/s41577-023-00855-z. PMID: 36914822; PMCID: PMC10009822.
- von Kaisenberg CS, Brand-Saberi B, Christ B, Vallian S, Farzaneh F, Nicolaides KH. Collagen type VI gene expression in the skin of trisomy 21 fetuses. Obstet Gynecol. 1998 Mar;91(3):319-23. doi: 10.1016/s0029-7844(97)00697-2. PMID: 9491853.
- Quarello E, Guimiot F, Moalic JM, Simoneau M, Ville Y, Delezoide AL. Quantitative evaluation of collagen type VI and SOD gene expression in the nuchal skin of human fetuses with trisomy 21. Prenat Diagn. 2007 Oct;27(10):926-31. doi: 10.1002/pd.1803. PMID: 17602442.
- Weyer K, Glerup S. Placental regulation of peptide hormone and growth factor activity by proMBP. Biol Reprod. 2011 Jun;84(6):1077-86. doi: 10.1095/biolreprod.110.090209. Epub 2011 Jan 26. PMID: 21270431.
- Conti A, Fabbrini F, D'Agostino P, Negri R, Greco D, Genesio R, D'Armiento M, Olla C, Paladini D, Zannini M, Nitsch L. Altered expression of mitochondrial and extracellular matrix genes in the heart of human fetuses with chromosome 21 trisomy. BMC Genomics. 2007 Aug 7;8:268. doi: 10.1186/1471-2164-8-268. PMID: 17683628; PMCID: PMC1964766.
- Mollo N, Aurilia M, Scognamiglio R, Zerillo L, Cicatiello R, Bonfiglio F, Pagano P, Paladino S, Conti A, Nitsch L, Izzo A. Overexpression of the Hsa21 Transcription Factor RUNX1 Modulates the Extracellular Matrix in Trisomy 21 Cells. Front Genet. 2022 Mar 10;13:824922. doi: 10.3389/fgene.2022.824922. PMID: 35356434; PMCID: PMC8960062.
- Wilber A, Nienhuis AW, Persons DA. Transcriptional regulation of fetal to adult hemoglobin switching: new therapeutic opportunities. Blood. 2011 Apr 14;117(15):3945-53. doi: 10.1182/blood-2010-11-316893. Epub 2011 Feb 14. PMID: 21321359; PMCID: PMC3087525.
- Zorick TS, Mustacchi Z, Bando SY, Zatz M, Moreira-Filho CA, Olsen B, Passos-Bueno MR. High serum endostatin levels in Down syndrome: implications for improved treatment and prevention of solid tumours. Eur J Hum Genet. 2001 Nov;9(11):811-4. doi: 10.1038/sj.ejhg.5200721. PMID: 11781696.
- Caglayan AO, Baranoski JF, Aktar F, Han W, Tuysuz B, Guzel A, Guclu B, Kaymakcalan H, Aktekin B, Akgumus GT, Murray PB, Erson-Omay EZ, Caglar C, Bakircioglu M, Sakalar YB, Guzel E, Demir N, Tuncer O, Senturk S, Ekici B, Minja FJ, Šestan N, Yasuno K, Bilguvar K, Caksen H, Gunel M. Brain malformations associated with Knobloch syndrome--review of literature, expanding clinical spectrum, and identification of novel mutations. Pediatr Neurol. 2014 Dec;51(6):806-813.e8. doi: 10.1016/j.pediatrneurol.2014.08.025. Epub 2014 Sep 4. PMID: 25456301; PMCID: PMC5056964.
- Palminiello S, Kida E, Kaur K, Walus M, Wisniewski KE, Wierzba-Bobrowicz T, Rabe A, Albertini G, Golabek AA. Increased levels of carbonic anhydrase II in the developing Down syndrome brain. Brain Res. 2008 Jan 23;1190:193-205. doi: 10.1016/j.brainres.2007.11.023. Epub 2007 Nov 22. PMID: 18083150.
- Haseeb A, Huynh E, ElSheikh RH, ElHawary AS, Scelfo C, Ledoux DM et al. Down syndrome: a review of ocular manifestations. Ther Adv Ophthalmol. 2022;30;14:25158414221101718.
- Palminiello S, Kida E, Kaur K, Walus M, Wisniewski KE, Wierzba-Bobrowicz T, Rabe A, Albertini G, Golabek AA. Increased levels of carbonic anhydrase II in the developing Down syndrome brain. Brain Res. 2008 Jan 23;1190:193-205. doi: 10.1016/j.brainres.2007.11.023. Epub 2007 Nov 22. PMID: 18083150.
- Ryan C, Vellody K, Belazarian L, Rork JF. Dermatologic conditions in Down syndrome. Pediatr Dermatol. 2021 Nov;38 Suppl 2:49-57. doi: 10.1111/pde.14731. Epub 2021 Aug 21. PMID: 34418156.
- Choi JK. Hematopoietic disorders in Down syndrome. Int J Clin Exp Pathol. 2008 Jan 1;1(5):387-95. PMID: 18787621; PMCID: PMC2480572.
- Bordon Y. Immune dysregulation in Down syndrome. Nat Rev Immunol. 2023 Apr;23(4):201. doi: 10.1038/s41577-023-00855-z. PMID: 36914822; PMCID: PMC10009822.
- Veteleanu A, Pape S, Davies K, Kodosaki E, Hye A, Zelek WM, Strydom A, Morgan BP. Complement dysregulation and Alzheimer's disease in Down syndrome. Alzheimers Dement. 2023 Apr;19(4):1383-1392. doi: 10.1002/alz.12799. Epub 2022 Sep 23. PMID: 36149090; PMCID: PMC10798358.
- Sullivan KD, Evans D, Pandey A, Hraha TH, Smith KP, Markham N, Rachubinski AL, Wolter-Warmerdam K, Hickey F, Espinosa JM, Blumenthal T. Trisomy 21 causes changes in the circulating proteome indicative of chronic autoinflammation. Sci Rep. 2017 Nov 1;7(1):14818. doi: 10.1038/s41598-017-13858-3. PMID: 29093484; PMCID: PMC5665944.
- Fatoba O, Itokazu T, Yamashita T. Complement cascade functions during brain development and neurodegeneration. FEBS J. 2022 Apr;289(8):2085-2109. doi: 10.1111/febs.15772. Epub 2021 Mar 1. PMID: 33599083.
- Pekna M, Pekny M. The Complement System: A Powerful Modulator and Effector of Astrocyte Function in the Healthy and Diseased Central Nervous System. Cells. 2021 Jul 17;10(7):1812. doi: 10.3390/cells10071812. PMID: 34359981; PMCID: PMC8303424.
- Grimm J, Heckl D, Klusmann JH. Molecular Mechanisms of the Genetic Predisposition to Acute Megakaryoblastic Leukemia in Infants With Down Syndrome. Front Oncol. 2021 Mar 11;11:636633. doi: 10.3389/fonc.2021.636633. PMID: 33777792; PMCID: PMC7992977.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.