Biology Group . 2023 March 22;4(3):485-501. doi: 10.37871/jbres1701.
Microbial Community Associated with Ground waters Discharge in Transylvania (Romania) and Balaton Highland (Hungary)
Marwene Toumi1*, István Máthé2, Ádám Tóth3, Nóra Sztráda1, Rózsa Farkas1, Laura Jurecska1 and Erika Tóth1
2Department of Bioengeneering, Faculty of Economics, Socio-Human Sciences and Engineering, Sapientia Hungarian University of Transylvania, 530104 Miercurea Ciuc, Libertății Sq. nr. 1, Romania
3József & Erzsébet Tóth Endowed Hydrogeology Chair, Department of Geology, Institute of Geography and Earth Sciences, ELTE Eötvös Loránd University, H-1117, Budapest, Pázmány P. stny 1/C, Hungary
- Microbial community
- Diversity
- Nutrient depleted aquatic environments
- NGS
- Total cell counts
- Taxon-specific
- Multitemplate PCR
Abstract
In the present study, water samples from 8 sites known as: Taploca, Nagy-borvíz, Piricske (located in Romania) and Szent Jakab, Kiskút, Kossuth Lajos, Polányi kút and Berzsenyi (located in Balaton highland in Hungary) and characterized with low nutrient content, were studied using cultivation independent methods. Diversity indices and cell counts were determined to assess the species richness in relation to the cell counts within the samples. Next generation sequencing was used to reveal the existing microbial community, and taxon specific PCR was used to detect the presence of some species with hygienic. 18 bacterial phyla above a ratio of 2% were identified in addition to 13 archaeal phyla using amplicon sequencing.
Nagy-borvíz water sample showed a unique presence of Chloroflexi, Bacteroidota, Desulfobacterota, Acidobacteriota and Actinobacteriota. Piricske water sample showed mostly Caldisericota and Spirochaetota. Taploca water sample contained a unique presence of Planctomycetota and Myxococcota at the same time with Szent Jakab water sample which showed an additional presence of Chloroflexi, Berzsenyi contained Campylobacterota at the same time with Polányi kút which had important fractions of Nitrospirota and Desulfobacterota. Finally, Kossuth Lajos water sample was dominated by Fusobacteriota, mainly members of Hypnocyclicus genus.
Piricske and especially Nagy-borvíz water samples contained important fractions of Altiarchaeota, moreover, Piricske contained high fraction of Euryarchaeota phylum -most of them belong to Methanobacteriaceae family.
Nitrosopumilaceae dominated Taploca and Kiskút in addition to their important presence in the rest of the samples. Diversity indices and cell count showed in general that cell count values are tending to be lower when the diversity is higher. The results of taxon specific PCR reactions showed that water treatment would be essential in some water samples.
Introduction
Recently, natural mineral water consumption has risen all over the world, becoming an alternative for tap water in many countries [1], Most of these waters are taken from naturally discharging springs or groundwater wells. These waters represent complex ecosystems with high diversity of microorganisms [2]. Taking into consideration that the European Union directive, restrains any water treatments aiming the removal or destruction of the autochthonous microorganisms, natural mineral waters are distributed containing the original microorganisms of the sources [2].
While distributing natural mineral water with its original microorganisms from the source can have some benefits such like keeping the microbial diversity and the water quality, there are also potential risks that need to be considered.
These risks include:
Microbial contamination: Natural mineral water can contain microorganisms, including bacteria and viruses that may pose a risk to human health.
Unpredictable microbial activity: The microbial activity can be unpredictable, and certain microorganisms may become dominant under other conditions. This can lead to changes in the water quality, which may not be desirable for consumers.
The identification and characterization of the autochthonous microbial community of the ground waters is of great importance for several reasons:
Understanding microbial ecology: Groundwater microbial communities play a crucial role in biogeochemical cycling of nutrients, such as carbon, nitrogen, and sulfur. The identification and characterization of these communities help us understand the ecological roles and functions of different microbial groups and their interactions with each other and the environment.
Monitoring environmental changes: Groundwater microbial communities are highly sensitive to changes in environmental conditions, such as changes in temperature, pH, and nutrient availability. Monitoring changes in these communities can provide valuable information about changes in the groundwater ecosystem and can help us identify potential environmental threats or contamination. The aim of the present study was performed on samples collected from groundwaters.
Three of the analysed springs are issuing from carbonate groundwaters located in the southern part of Harghita County, Romania. This region is characterized with an average temperatures that fall between 20 degrees celsius (68°F) and 25 degrees celsius (77°F) during the month of June, July, August and September. The coldest month is January with an average maximum temperature of -2°C (27°F). The precipitation amounts to 595 mm/year.
The other five investigated springs are situated in Balaton Highland region, this area is characterized with a humid continental climate, but the effects of the Mediterranean influence can be detected in the sunshine duration and annual mean temperature (11°C). The amount of precipitation, which falls predominantly in the form of rain is 500 to 600 mm/year. The Balaton Highland is the most extensive carbonate aquifer system in Hungary. The region is abundant in naturally discharging springs of various temperatures, creeks and wetlands.
The aim of this study is to: 1). Check the microbial cell counts and diversity indices in different ground waters. 2). Reveal the microbial community structure in these systems using cultivation independent method (NGS) and 3: determine the risks and possible antibiotic resistance among the detected bacteria.
Materials and Methods
Description of the hydrogeological properties of the sampling sites
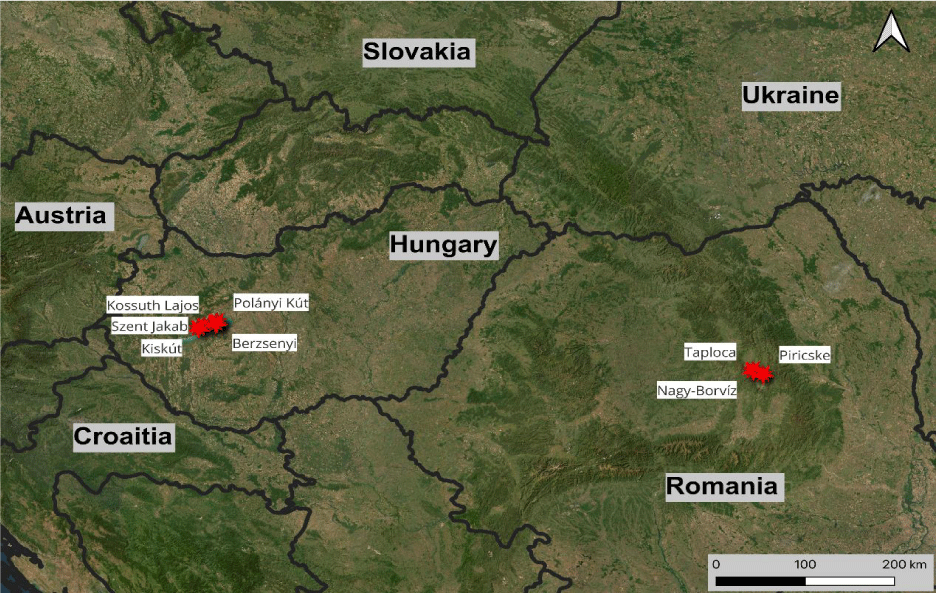
Three of the analyzed springs are located in the southern part of Harghita county, Romania: Taploca spring (46.3697°N, 25.8055°E) is located in the Csíktaploca (Toplița Ciuc), Nagy-borvíz spring (46.3753°N, 25.8193°E) (borvíz [“wine water”] means CO2 rich mineral water) is located in Csíksomlyó (Şumuleu Ciuc) near the pilgrimage site. Today Csíktaploca and Csíksomlyó villages are part of the Csíkszereda (Miercurea Ciuc) town. Piricske spring (46.3696°N, 25.7954°E) is found in the forest close to Csíkszereda. Piricske is a freshwater spring. Taploca and Nagy-borvíz springs are CO2- rich mineral waters, their dominant ions are HCO3–, Na+, Ca2+, Mg2+ and Fe2+ [3]. The hydrochemical character of these discharging groundwater is influenced and determined by the geological structure and groundwater-rock interaction resulting in high mineralization of NaCl, sulfate and dissolved gases (CO2, H2S) [4]. Five of the investigated springs are situated in Balaton Highland region: Szent Jakab spring of Vászoly (46.9433°N, 17.7580°E) and Kiskút of Szentantalfa (46.9126°N, 17.6745°E) are located in the elevated hills (~280 and ~190 m above sea level) and the Kossuth Lajos (46.9561°N, 17.8950°E), Polányi kút (46.9434°N, 17.8669°E) (Szekér Ernő outflow) and Berzsenyi spring (46.9465°N, 17.8739°E), can be found in Balatonfüred near the shoreline of the Lake Balaton (~105-115 m above the sea level). These springs issue from carbonate aquifers or at the boundary of sandstone, metamorphic and carbonate formations, where a hydraulic barrier forces groundwater discharge [5]. The water temperature in the elevated parts is close to the mean annual temperature of the air (~10-11°C) because these springs are fed by local and shallow groundwater flow. In turn, the springs of Balatonfüred receives a deeper groundwater flow component which is responsible for the higher water temperature (~14-17°C) and the natural occurrence of the CO2 [6]. The figure bellow is showing the location of the different samples.
Collection of water sample
The water sample located in Romania (Taploca, Nagy-borvíz, Piricske) and in Balaton Highland region (Szent Jakab, Kiskút, Kossuth Lajos, Polányi kút and Berzsenyi) were collected from the spring’s water outflow. The sampling was carried out during the period from 2018 to 2021. The water samples (2-2 L) were aseptically collected into clean, sterile, glass bottles according to ISO 19458:2006 standard, transferred at 4TFFC in a cooler bag and filtered for cell count determination and molecular studies immediately upon arrival at the laboratory.
Determination of the physical and chemical parameters of the samples
The physical and chemical parameters of the samples were determined by measuring each sample in replicates to ensure accuracy and precision.
The pH and temperature were measured on site using a Hach HQ40D portable multimeter (Hach, Loveland, CO, USA). All other parameters mentioned bellow were determined in the laboratory according to standard methods [7]. Nitrite ion (ASTM 4500-NO2--B) was measured following the colorimetric method, using Hach DR2000 spectrophotometer (Loveland, Colorado, USA). Nitrate ion (ASTM 4500-NO3--B) was measured using the Ultraviolet Spectrophotometric Screening Method, using Perkin Elmer Lambda 35 UV/VIS spectrophotometer (Waltham, Massachusetts, USA). Sulfate ion (ASTM 4500-SO42--E) and iron (3500-Fe-D) were measured with Hach DR2000 spectrophotometer (Loveland, Colorado, USA). The amount of total organic carbon (TOC, ASTM 5310-B) Combustion-Infrared Method was determined by a Multi N/C 2100S analyser (Analytik Jena, Germany). Ortophosphate (ASTM 4500-P-E) was measured following the Ascorbic Acid Method, Hach DR2000 spectrophotometer (Loveland, Colorado, USA).
Determination of the total cell counts of the samples
200 ml from each replicate of each water sample was filtered using a polycarbonate membrane filter (0.2 μm GTTP, Millipore, USA), the obtained filters were treated with 2% paraformaldehyde solution (Sigma-Aldrich, Germany) dissolved in 0.1 m phosphate buffer (NaH2PO4 3.2 g, Na2HPO4 10.9 g in 1000 ml distilled water, pH 7.2) overnight in order to fix the cells. The obtained filters were stored at -20°C until further analysis. Microscopic cell counts were determined using Nikon80i epifluorescent microscopy and NisElements program package according to [8].
The images were analyzed using the image analysis software to count the cells in each image, and the mean cell count was calculated across the 20 images.
DNA extraction from the water samples and amplicon sequencing
For DNA extraction, 250 ml water sample from each replicate was filtered using a 0.22 μm pore size sterile mixed cellulose filter (MF-Millipore GSWP04700, Billerica, MA, USA) using DNeasy® PowerSoil® DNA Isolation Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions [9].
A mechanical cell disruption was performed by shaking the filters at 30 Hz for 2 min using a Retsch Mixer Mill MM400 (Retsch, Haan, Germany).
To perform the PCR reactions 3 μl quantity of the template DNA was used. The PCR reaction for amplifying the 16S V4 region was done in triplicate based on the following protocol: 98°C for 3 min; 25 cycles: 95°C for 30s, 55°C for 30s and 72°C for 30s; and 72°C for 5 min for bacteria and 98°C for 3 min; 25 cycles: 95°C for 30s, 60°C for 30s and 72°C for 30s; and 72°C for 10 min for archaea using the following primers: CS1-TS-B341F and CS2-TS-805NR for bacteria [10] and CS2-TS-Arch-855R and CS1-F-A519F for archaea [11]. Before sequencing, DNA concentration of the PCR products was determined using a Qubit meter (Invitrogen life technologies corporation, Austria) and a minimal concentration of 4 ng/μl and 50μl of PCR product was respected.
Illumina MiSeq platform with MiSeq standard v2 chemistry was used by the Genomics Core Facility RTSF, Michigan State University to perform the amplicon sequencing.
The forward and reverse fastq files obtained from Illumina sequencer, were processed and analyzed using the Mothur v1.41.5 software [12] following the protocol of Benedek [13]. Deltaq was adjusted to 10 with the command “make.contigs”, for chimera detection UCHIME [14] was used within the mothur commands, singletons were removed according to Kunin [15].
The sequences taxonomy were classified using he ARB-SILVA SSU Ref NR 138_1 reference database [16], bootstrap confidence score is more than 80 with 1000 iterations. Based on the taxonomic classification output, both non-archaeal and non-bacterial sequences were removed from the analyses.
The Operational Taxonomic Units (OTUs) were determined using a distance matrix with distances larger than 0.3 [17].
Eventually, the Mothur’s commands, sub sample was used to normalize the data, in fact reads were subsampled to the read number of the sample having the lowest sequences count.
The command rarefaction single was used to calculate the rarefaction curve in addition to Shannon-Weaver and inverse Simpson (1/D) diversity indices and Chao-1 and ACE (Abundance-based Coverage Estimator) richness metrics.
Sequence reads were deposited in the NCBI SRA database and are accessible through the Bio Project ID PRJNA628507, under the references: SRS6537389 for Taploca, SRS6537391 for Piricske, SRS6537423 for Nagy-borvíz, SRS9983636 for Berzsenyi, SRS9983634 for Polányi kút, SRS9983633 for Kossuth Lajos, SRS9983477 for Kiskút and SRS9983632 for Szent Jakab.
Taxon specific and multitemplate PCR reactions
The presence of the following bacterial groups was tested using taxon specific/multitemplate PCR reactions, using the same DNA extracted for NGS analysis: coliform bacteria and E. coli according to Bej AK, et al. [18]; Acinetobacter baumannii based on Huang LY, et al. [19] Legionella sp. [20]. Stenotrophomonas maltophilia [21]; Legionella pneumophila [22]. Pseudomonas aeruginosa according to Lavenir R, et al. [23]. The ESBL producing bacteria were tested by multiplex PCR according to Trung NT, et al. [24] and the presence of macrolide resistance genes by simultaneous detection of 5 genes (ermA, ermB, ermC, msrA, mef) according to Zmantar T, et al. [25].
Statistical analysis
Environmental variables (physical and chemical parameters), diversity indices, cell counts and the revealed OTUs (archaea and bacteria) were represented by Principal Components Analysis Ordination (PCA) put together with vector-fitting [26].
In order to fit the variable as vectors, the “envfit” function from the vegan package was applied [27].
Shannon diversity index was calculated in order to assess the population diversity within the samples based on Operational Taxonomic Units (OTUs) [28].
Results
Physical and chemical parameters of the water samples
Physical and chemical parameters of the different water samples are given in table 1.
Microscopic cell counts and diversity indices of the samples
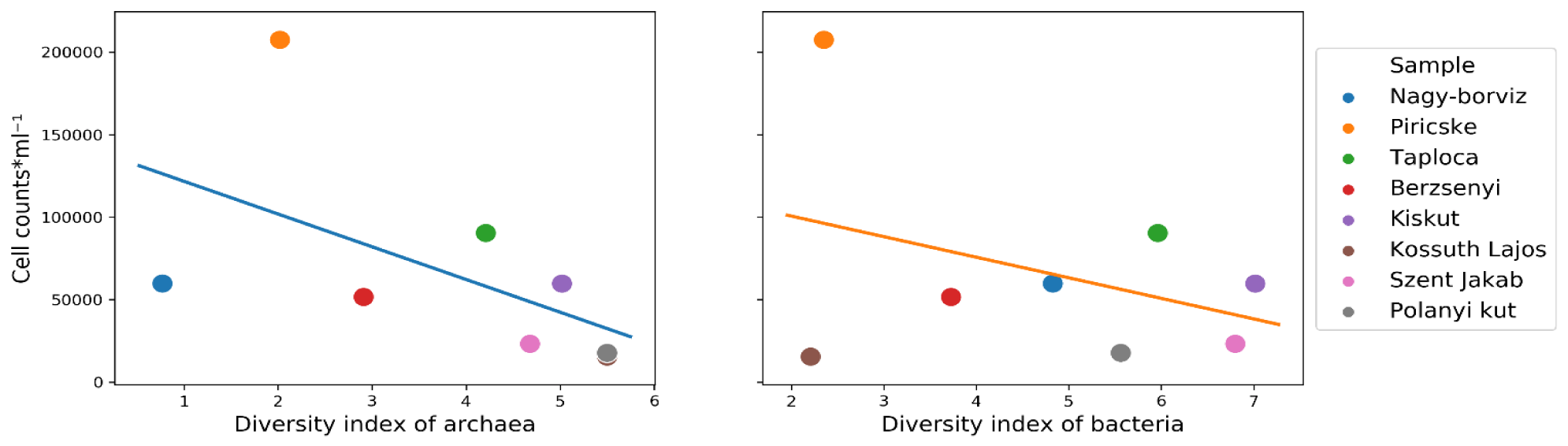
The relationship between the number of bacteria (cell count values) and diversity among the samples is shown on figure 1. The cell count values of Piricske water sample were higher than all the other samples by one order of magnitude (106) (Table 1). The majority of the samples had similar Shannon diversity index values between archaea and bacteria. The exception was detected in the case of Kossuth Lajos water sample where it was the most diverse in term of archaea and the least diverse in case of bacteria, the opposite was observed in the case of Nagy-borvíz water sample. A general trend was seen within the majority of the samples (except Kossuth Lajos) showing that the cell count values is tending to be lower when the diversity is higher (Figure 2) (Supplementary Tables 1,2).
Bacterial community composition of the different samples based on amplicon sequencing
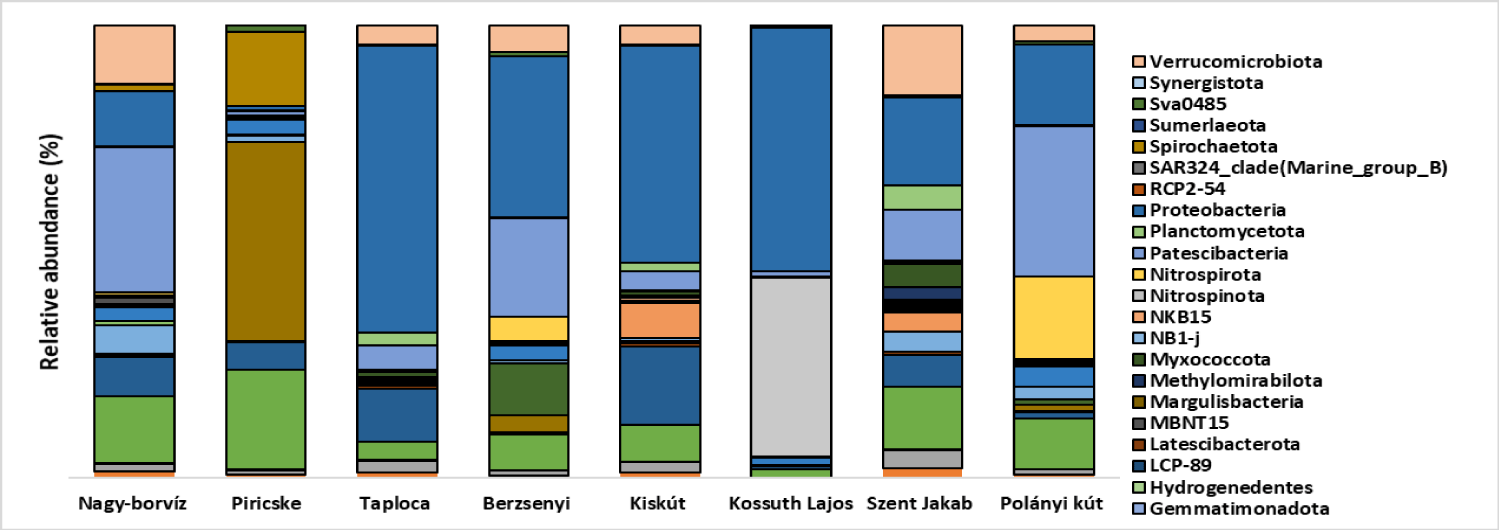
The results of amplicon sequencing identified 18 bacterial phyla presented within a ratio higher than 2% in at least one of the 8 samples. The results of the rarefaction curves (Supplementary figure 1) showed that the sequencing depth was sufficient to identify the majority of the bacterial taxa.
Verrucomicrobia phylum was present in all samples with high abundance (ranging from 3% to 15%) except Piricske water sample. Most of the identified members of this phylum belongs to the Omnitrophaceae family.
All samples had distinct bacterial community. In fact, Nagy-borvíz water sample showed the presence of remarkable fractions of Chloroflexi, Bacteroidota, Desulfobacterota, Acidobacteriota and Actinobacteriota. Piricske water sample was dominated by Caldisericota, also showed the presence of important fraction of Spirochaetota. Taploca water sample contained a unique presence of Planctomycetota and Myxococcota at the same time with Szent Jakab water sample which showed an additional presence of Chloroflexi, Cyanobacteria, Bacteroidota and Actinobacteriota. Berzsenyi contained Campylobacterota at the same time with Polányi kút which had important fractions of Nitrospirota and Desulfobacterota. Finally, Kossuth Lajos water sample was dominated by Fusobacteriota which contained mainly members of Hypnocyclicus genus.
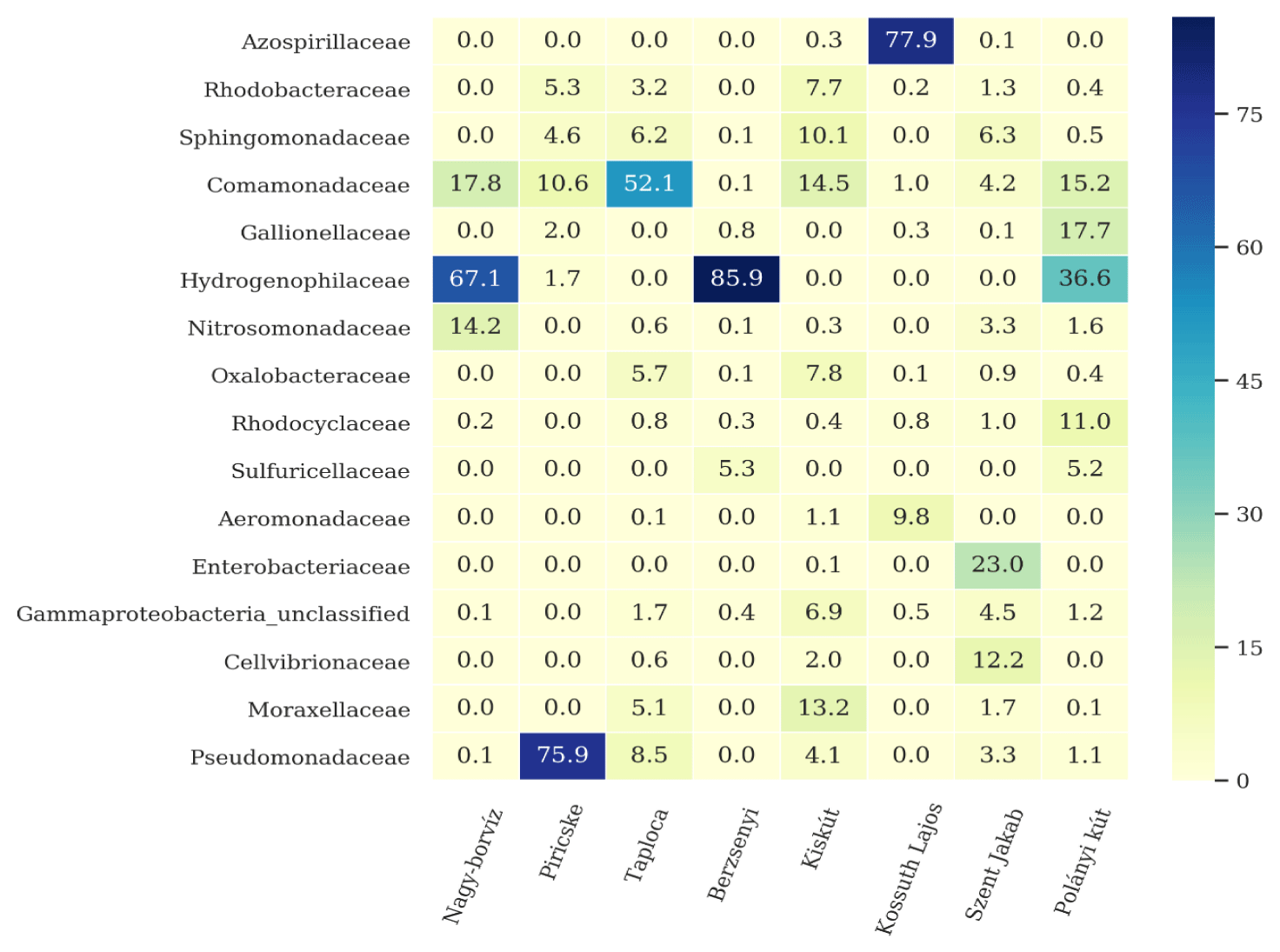
All samples were characterized by the abundance of Proteobacteria (Figure 3), and Patescibacteria. The phylum Patescibacteria contained high ratio of Parcubacteria and many candidatus genera like Falkowbacteria, Magasanikbacteria, Azambacteria, etc. Within the Proteobacteria (Figure 4). Nagy-borvíz water sample showed the presence of the families Comamonadaceae, Hydrogenophilaceae and Nitrosomonadaceae, Piricske water sample contained mainly members of the families Pseudomonadaceae and Spirochaetota, Taploca water sample is characterized by a high presence of the Caulobacteraceae family. Berzsenyi water sample contained mostly Hydrogenophilaceae and Sulfuricellaceae. Kiskút water sample contained unclassified both Alphaproteobacteria and Burkholderiales, Rhizobiaceae, Sphingomonadaceae, and Moraxellaceae. The Proteobacteria community of Kossuth Lajos water sample was split mainly between Aeromonadaceae and Azospirillaceae. Szent Jakab was not dominated by any family - similarly to Kiskút water sample - because of the high diversity within the Proteobacteria phylum in these samples, however, important fractions of Cellvibrionaceae and Enterobacteriaceae were identified, the latter family contained members of the genera Lelliottia and unclassified Enterobacteriaceae. Polányi kút contained a relatively important fraction of the three families Comamonadaceae, Gallionellaceae and Hydrogenophilaceae.
Archaeal community composition of the different samples based on amplicon sequencing
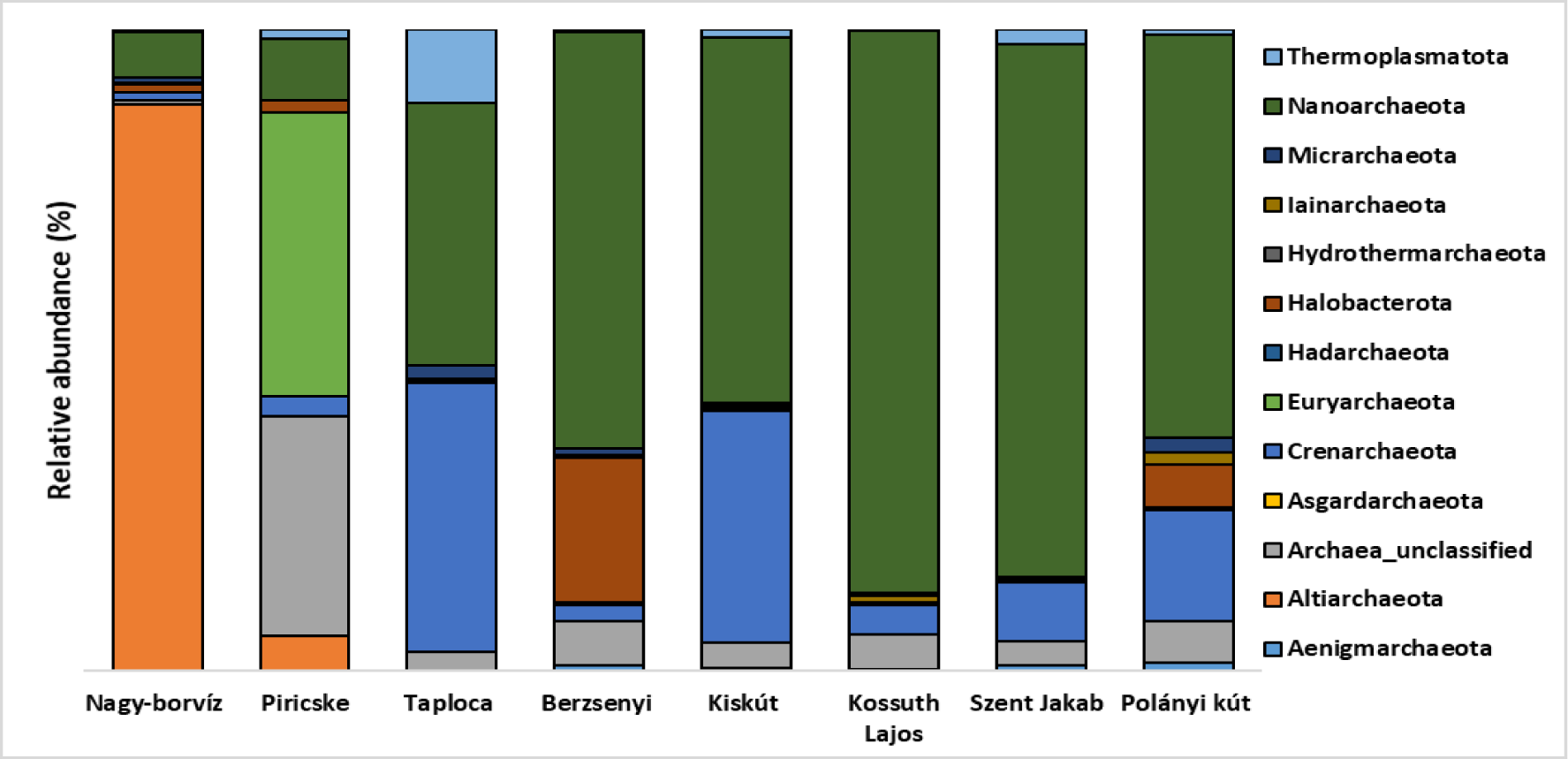
Altogether, 13 archaeal phyla were detected in the water samples (Figure 5). The rarefaction curves of the samples (Supplementary figure 2) showed that the sequencing depth was sufficient to recover the majority of the archaeal taxa.
Unclassified Archaea, Nanoarchaeota-belonging to the Woesearchaeales order -, Thermoplasmatota, Halobacterota - low fraction within Taploca water sample - and Crenarchaeota phyla were present in all the samples.
Thermoplasmatota phylum contained mainly unclassified families characterizing both Nagy-borvíz and Piricske water samples, the rest of the samples contained unclassified Thermoplasmata. Concerning Crenarchaeota phylum Nagy-borvíz and Piricske water samples contained mostly unclassified Crenarchaeota in addition to Bathyarchaeia taxa in case of Nagy-borvíz. The rest of the samples contained variable fractions of taxa belonging to the Nitrososphaeria class.
Piricske and especially Nagy-borvíz water samples contained important fractions of Altiarchaeota, moreover, Piricske contained higher fraction of Euryarchaeota phylum-most of them belong to Methanobacteriaceae family. With the exception of Piricske water sample, all the rest of the samples contained variable fractions of Micrarchaeota.
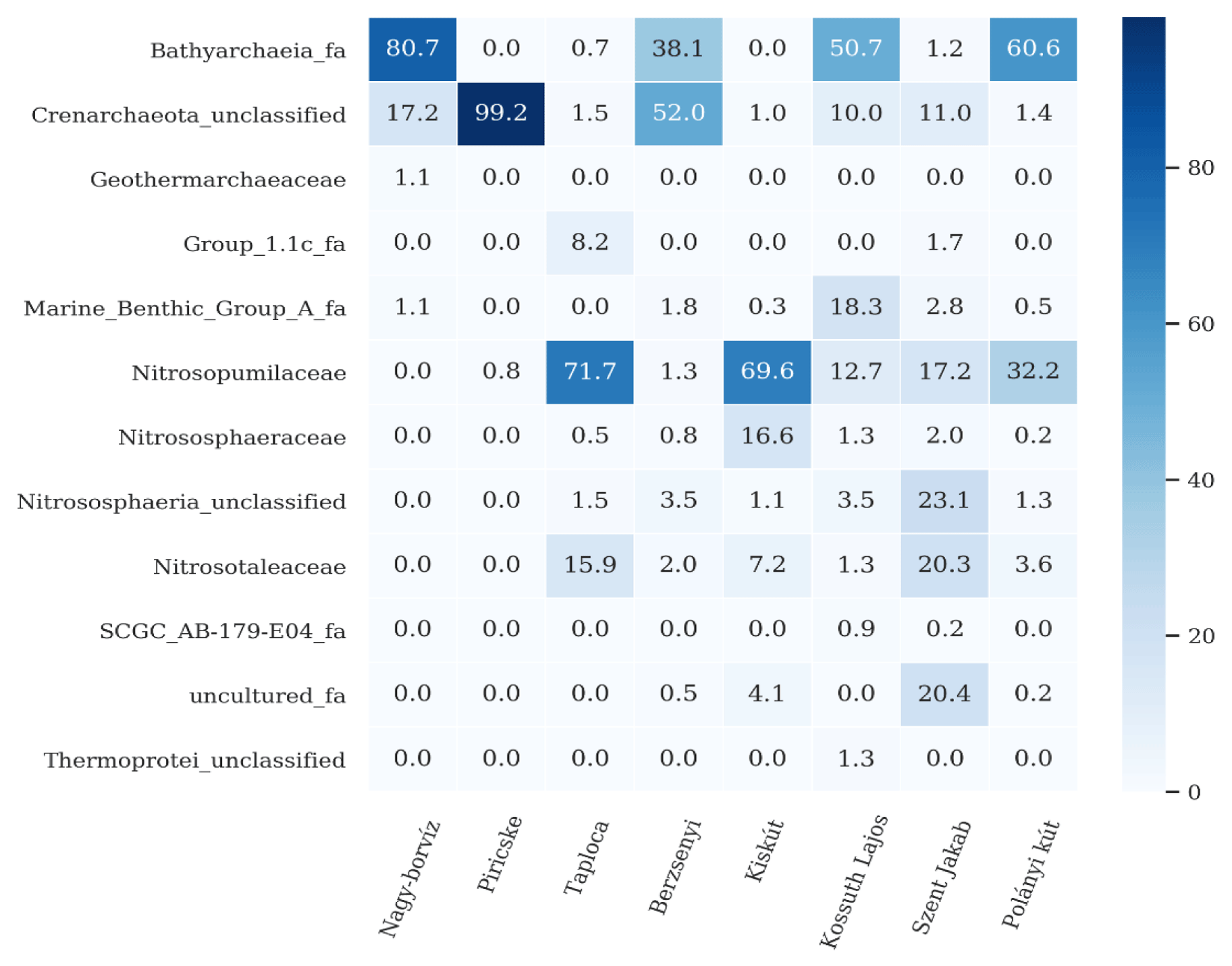
Among the families in the Crenarchaeota phylum, all samples contained the following ammonia oxidizing archaea (figure 6) except Nagy-borvíz and Piricske water samples:
Nitrosopumilaceae: dominated Taploca and Kiskút with important presence in the rest of the samples, Nitrososphaeraceae is mainly present in Kiskút, unclassified Nitrososphaeria is present with important fraction in Szent Jakab. Lastly, Nitrosotaleaceae is present with important fractions in Taploca, Kiskút and Szent Jakab.
Results of taxon-specific and multitemplate PCR reactions
The genus Legionella was present in all the samples except Piricske water sample, but not in any case L. pneumophila was detected. Pseudomonas aeruginosa, E. coli or coliform bacteria were not present in the samples. The presence of Stenotrophomonas maltophilia (being an emerging facultative pathogen bacterium) which is a potential cause of concern, was present in six of the samples. On the other hand, neither ESBL producer, nor macrolide resistant bacteria were detected (Table 2).
Discussion
Microbial communities of the different samples based on amplicon sequencing
According to the results of Kuznetsov SI, et al. [29], oligotrophic environments are characterized by less than 1.36 mg/L suspended organic substance and 15.24 mg/L for dissolved organic material. More recent studies such like the one obtained by Ho A, et al. [30] confirmed that oligotrophs are characterized by their ability to grow under low substrate concentrations (e.g. carbon in the nano and picomolar range).
TOC values of our samples are lower in each case therefore all samples can be stated oligotrophic.
All samples were characterized by a high percentage of unclassified and uncultured OTUs, table 3 and table 4 further demonstrate this large fraction, in fact over 27% in case both Archaea and Bacteria were not classified with any specific taxa and were therefore determined as Unclassified organisms.
| Table 3: Top 2 archaea taxa affiliations. Known taxa affiliations from OTU taxon classification were condensed to display the percentage of that taxon in the dataset. | |
| Taxonomy | % Across Dataset |
| Archaea; Nanoarchaeota; Nanoarchaeia; Woesearchaeales; Woesearchaeales_unclassified; Woesearchaeales_unclassified | 15.91 |
| Archaea; Archaea_unclassified; Archaea_unclassified; Archaea_unclassified; Archaea_unclassified; Archaea_unclassified | 7.91 |
| Table 4: Top 2 bacteria taxa affiliations. Known taxa affiliations from OTU taxon classification were condensed to display the percentage of that taxon in the dataset. | |
| Taxonomy | % Across Dataset |
| Bacteria; Bacteria_unclassified; Bacteria_unclassified; Bacteria_unclassified; Bacteria_unclassified; Bacteria_unclassified | 10.4 |
| Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Comamonadaceae; Comamonadaceae_unclassified | 1.84 |
Previous studies show that low nutrient content environments and groundwaters discharge data sets are usually dominated by unknown taxa. Previous studies of Lopez-Fernandez M, et al. [31] and Gayner NJ [32] showed that almost 50% of the identified phyla in groundwater samples were archaeal or bacterial candidates, moreover, the percentage of unknown and candidate phyla increased with depth, which highlights the importance of further studies to characterize deep biosphere microbial communities.
Even thought, a high percentage of OTUs affiliation are unclassified taxa, many of the classified ones are associated with previously known groundwater and poor nutrient environments microbes.
Many taxa are recently discovered as CPR and DPANN organisms.
Woesarchaeota, presented in a relatively large proportion of this microbial dataset (10%), especially in Szent Jakab and Kossuth Lajos water samples with a percentage of 16.92 % and 17.55%, respectively. They are part of DPANN superphylum, previous studies based on genome analyses and metabolic reconstructions propose that they are anaerobic heterotrophic organisms with a potential of symbiotic relationships especially with methanogens [33,34]. This feature can remove the thermodynamic bottlenecks and enables several metabolic reactions under nutrient-depleted conditions [35]. Some studies present Woesarchaeota as fermenters with iron or methane metabolism [36].
Amplicon sequencing showed the presence of members of Thermoplasmatota phylum, known previously of inhabiting not only extreme environments but also a wide range of environments [37]. Taxa existing within this phylum could be classified only in case of Nagy-borvíz and Piricske water samples where mainly they belonged to Marine_Benthic_Group_D (Thermoprofundales) and DHVEG-1 (Thermoplasmata).
Based on few available cultures and genomes of Thermoplasmata, researchers could have an insight on their myxotrophic lifestyle [38] which is crucial for their sustainability in oligotrophic ecosystems in order to compensate the lack of organic nutrients [39].
Shuming MCJ, et al. [40] demonstrated that Crenarchaeota and Halobacterota had relatively complete gene families involved in dissimilatory sulfate reduction, these findings are in accordance with the chemistry analyses as Taploca water sample (characterized with the lowest fraction of Halobacterota) contained the lowest amount of SO42-. Sulfate-Reducing Bacteria (SRB) and Sulfate-Reducing Archaea (SRA) can use sulfate as a terminal electron acceptor in anaerobic respiration. During this process, sulfate is reduced to sulfide, which can lead to the accumulation of hydrogen sulfide gas in the water. Hydrogen sulfide gas can be toxic to other aquatic microorganisms.
Among the families under the Crenarchaeota phylum, all the samples except Nagy-borvíz and Piricske water samples contained the following ammonia oxidizing archaea: Nitrosopumilaceae [41], Nitrososphaeraceae [42] and Nitrosotaleaceae [43] (Figure 6). These Archaea are responsible for the first step of nitrification, which is the oxidation of ammonia to nitrite. The produced nitrite is then oxidized by other microbes to nitrate, which can accumulate in the spring water.
Most of these samples contained higher values of NO3- (Table 1), this can be explained by the high rates of nitrification in which certain bacteria oxidize ammonia to nitrite, and then further oxidize nitrite to nitrate. This process can increase the nitrate concentration in spring water [44].
Nagy-borvíz water sample which is characterized by high ammonia content, contained members of Nitrosomonadaceae family which are lithoautotrophic ammonia oxidizing bacteria [45]. In addition, species from the phyla candidate division Zixibacteria-found especially in Piricske water sample and Chloroflexi-present in all the samples also known to contain nitrification genes [46].
Candidatus Parcubacteria, considered as a part of CPR superphylum, members of this phylum are known to harbour a variety of metabolisms with the possibility of acquiring fermentative processes able to produce acetate, ethanol, lactate, and hydrogen [32]. Previous genomic analyses of Parcubacteria revealed the existence of nitrite reductases which can transform nitrite to produce nitric oxide (gene nirK) and ammonium (gene nirB) [36].
The predominance of the superphylum Patescibacteria in groundwaters is often related to their mobilization from soils and their good survival under oligotrophic conditions [47]. Co-occurrence network analysis pointed to potential associations of Patescibacteria with specific organisms involved in nitrogen, sulfur and iron cycling [47]. Other capability of Candidatus Patescibacteria members in oligotrophic habitats is their abundance under ultra-small cells and acquirement of reduced genome size (pass through 0.2 µm pore size filter) [48,49]. These features are thought to be evolutionarily advantageous, as the increased surface-to-volume ratio optimizes the uptake of the sparse nutrients [50] and the loss of expendable genes leads to a lower metabolic cost of reproduction [51], these characteristic are also seen in Caldisericota genome [52] which was revealed in Piricske water sample.
Chloroflexi phylum known to harbour many phototrophic microorganisms - was present in all water samples. Members of this phylum are correlated with the fraction of TOC level within the environment, this explain their abundance in all the samples as they are characterized with TOC values below the oligotrophy level [53].
The rest of the OTUs were specific to one or many water samples, among them:
Altiarchaeaceae: High presence in Nagy-borvíz and Piricske water samples, literature shows that they predominate anaerobic groundwater due to evolved metabolic and structural features where they may represent an important carbon dioxide sink [54], cultivated member of this family has autotrophic metabolism [54].
Methanogenic archaea: They are present in many samples in different families, Methanobacteriaceae and Methanoregulaceae in Piricske, Methanoperedenaceae in Nagy-borvíz, Berzsenyi and Polányi kút water samples. They are able to thrive in nutrient-poor, low ionic-strength environments [55].
Sulfate-Reducing Bacteria (SRB): The phylum Desulfobacterota-found in most samples - contains sulfate-reducing and sulfate related fermentative and syntrophic taxa [56]. Cultured Desulfobacterota showed preference for anoxic conditions, and many utilize sulfate, sulfite, thiosulfate, elemental sulfur, or iron and amino acids) [60].
Members of the Sphingomonadaceae as terminal electron acceptor in respiratory and/or disproportionation processes [57].
The phylum Nitrospirota - found in many samples - currently includes a limited number of bacterial species with validly published names, among them Sulfate-Reducing Bacteria (SRB) [58], these taxa contain gene set for dissimilatory sulfate reduction [59].
Among proteobacteria phylum: Azospirillum genus dominated Kossuth Lajos water sample, it can use NH4+, NO3-, amino acids and N2 as nitrogen sources for growth. They can grow under anaerobic conditions using nitrate as electron acceptor, microaerobic (N2 or NH3 as nitrogen sources) and fully aerobic conditions with combined nitrogen only (NH3, NO3- family were detected in all the samples except Nagy-borvíz and Kossuth Lajos water samples (Table 5).
| Table 5: Most abundant members of the family Sphingomonadaceae within the samples and their potential ecological role. | ||
| Genus | Sample name | Description |
| Blastomonas | Berzsenyi Kiskút |
Can be isolated from oligotrophic environments [61], characterized with slow growing, this change in growth dynamics can increase the cell volume [62]. |
| Novosphingobium | Taploca Kiskút |
Known with high genomic and functional plasticity allowing it to rearrange its genome according to environment variations and adapt their metabolic capabilities to better cope with severe conditions (degrade a wide range of aromatic hydrocarbons) [63]. |
| Sphingomonas | Taploca Kiskút |
Has the ability to degrade the copper pipes in drinking water distribution systems (White, et al. 1996). |
| Sphingopyxis | Kiskút | Has the capacity to thrive in harsh environments [65], commonly isolated also from freshwater and marine habitats and many are facultatively chemolithotrophs, often producing H2 during their metabolism [33]. |
| Sphingorhabdus | Kiskút Szent Jakab |
Isolated from diverse environments [66] |
Iron oxidisers: Polányi kút water sample contained important ratio of the family Gallionellaceae where its members belong to Gallionella and Sideroxydans genera. Both are adapted to chemolithoautotropy [67]. Chemistry measurements showed important fraction of iron within that sample.
Hydrogenophilaceae: Most members of this family are chemolithotrophic using various inorganic electron donors such as reduced sulfuric compounds or hydrogen [32]. Sequences at the genus level could not be classified however, their high abundance in Nagy-borvíz, Polányi kút and Berzsenyi water samples - characterized with medium to high SO42- content comparing to the other samples can give an insight about their potential role as sulfate reducers. This is endorsed also by the presence of members of sulphur oxizing bacteria “sulfuricellaceae” in the last two mentioned samples.
Certain orders of Proteobacteria such as Burkholderiales, have been associated with nutrient poor conditions [68].
Influence of the environmental factors on the diversity of archaeal and bacterial communities.
It is known that hydrogeological and other ecological factors influence the microbiological processes [69]. On the other hand, the metabolism of microorganisms usually affects the water quality of groundwater systems [69]. In addition, the fate and transport of microorganisms in groundwaters are the result of their physicochemical characteristics (flow velocity, gain, size, porosity, solid organic carbon content, temperature, pH, and other chemical characteristics of the water) [70].
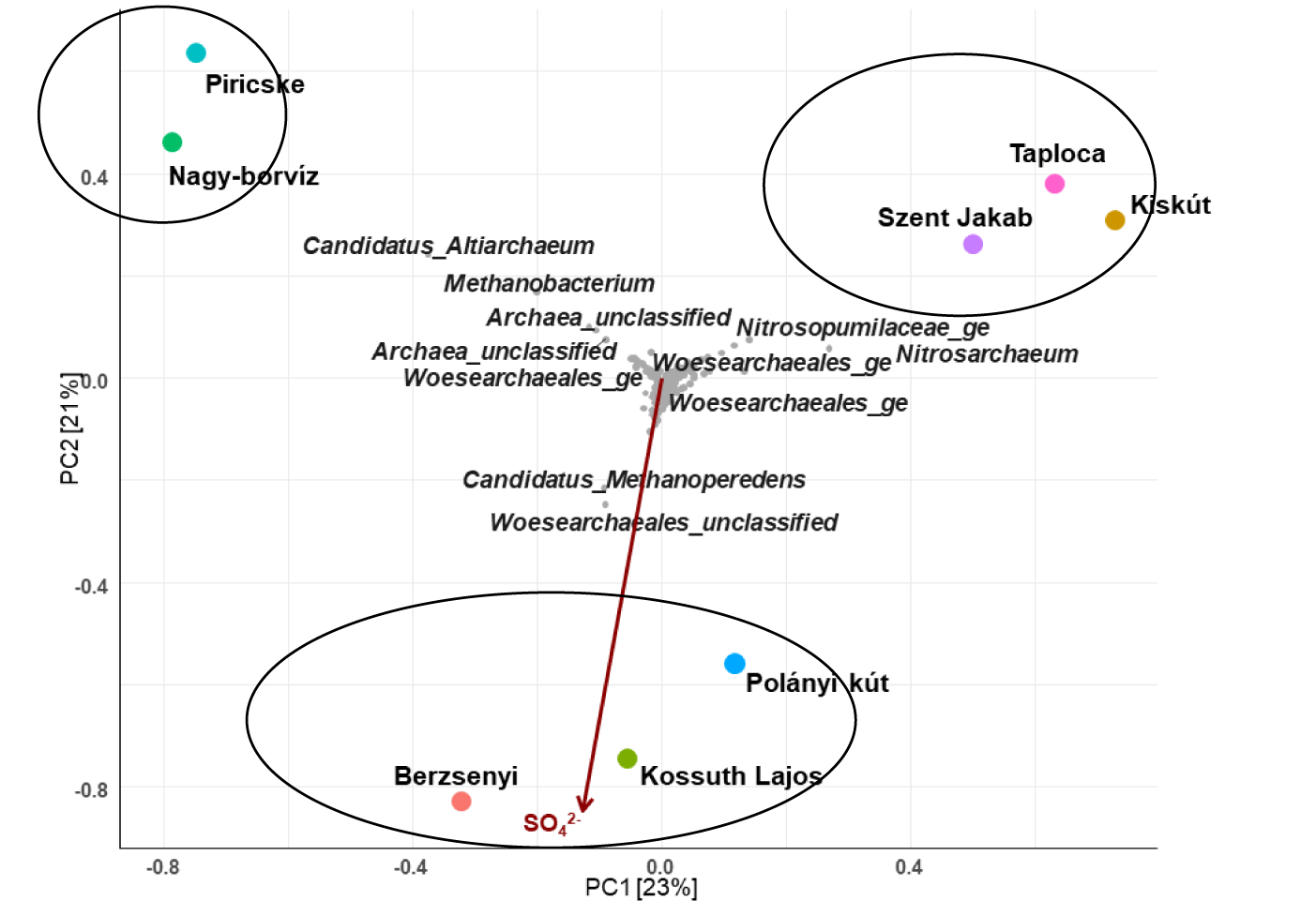
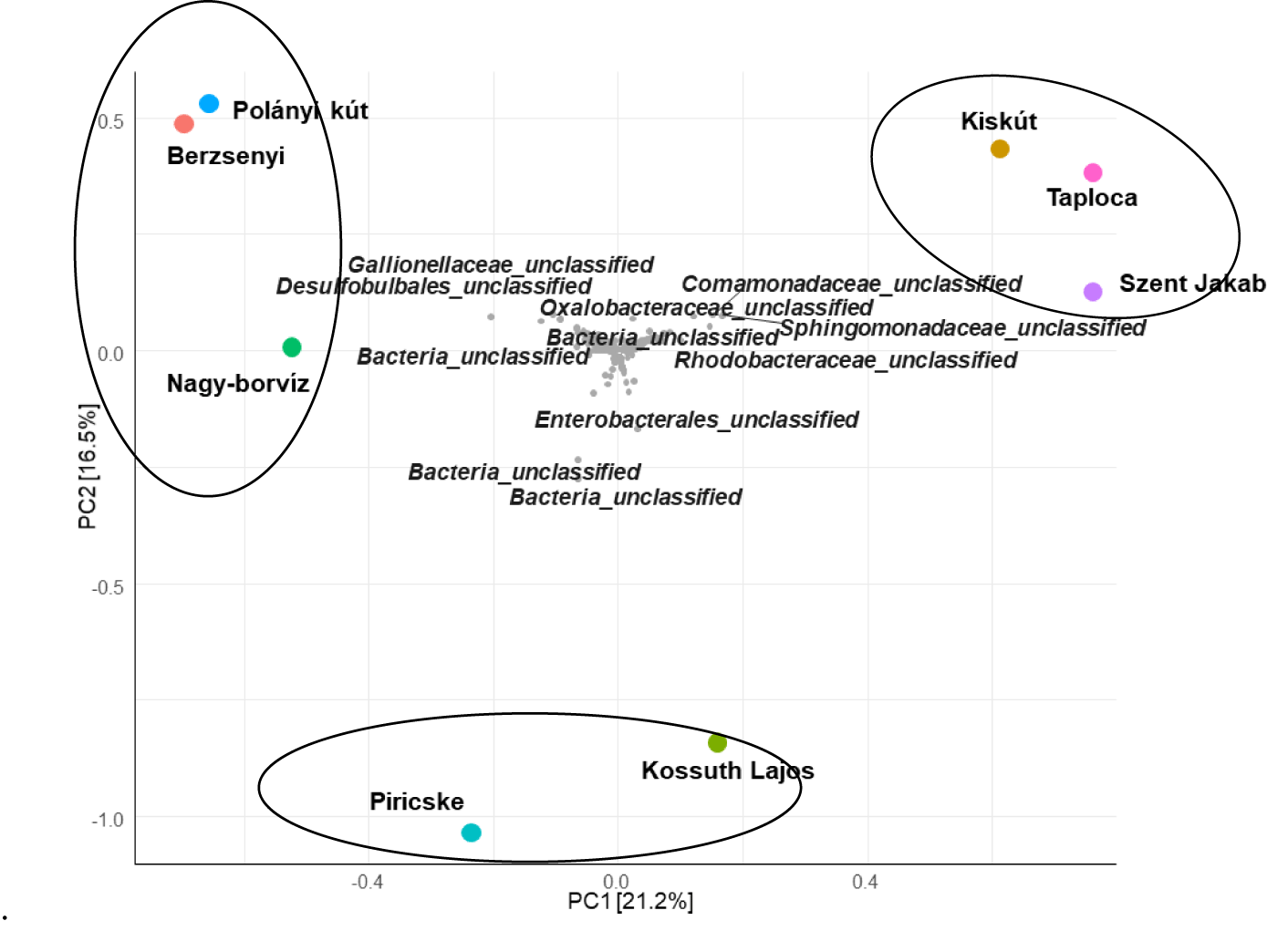
In order to assess the effect of environmental factors on the prokaryotic communities, the PCA ordination was performed (Figures 7,8). The analysis revealed the impact of chemical characteristics (p < 0.1) on archaea and bacteria OTUs distribution.
Microorganisms have an important role in mediating reactions in aquifers that affect geological processes, at the same time physical and chemical variations result in changes in microbial communities [71]. Nagy-borvíz and Piricske water samples were grouped together, they contained common presence of Altiarchaeum and Methanobacterium taxa (Figure 7). In previous studies, these two taxa appeared together in many other environments [72] where CO2 fixation could be carried out by members of the class Altiarchaeia meanwhile Methanobacterium utilize H2/CO2 as sole carbon and energy source. These two water samples are located in the Transylvanian region characterized with the high presence of dissolved gases (CO2, H2S).
Taploca, Szent Jakab and Kiskút water samples were grouped together due to the presence of OTUs related to the nitrogen cycle (as an example: Nitrosopumilaceae and Nitrosoarchaeum). These water samples contained the highest fractions of either NO3- or ammonia among the samples.
Sulfate had an important impact on the archaeal community structure of Berzsenyi, Kossuth Lajos and Polányi kút (Figure 7). The most abundant taxa in these samples is Methanoperedens (known previously to perform anaerobic oxidation of methane coupled to nitrate reduction [73]). The study of Timmers [74] showed that this taxon was found to be abundant in aquifers where methane and sulfate were present, moreover the absence of nitrate-dependent anaerobic oxidation of methane activity in the studied samples presented Methanoperedens as an anaerobic methane oxidizer coupled to Sulfate reduction.
Polány Kút, Berzseny and Nagy-borvíz water samples were related with the presence of Gallionellaceae and Desulfobulbales (Figure 8), they are known as ferrous iron-oxidizing and sulfate-reducing bacteria. These samples contained important fractions of iron (Table 1) allowing the abundancy and activity of these microbes in oxic/anoxic interfaces of sediments in groundwaters [75].
| Table 1: Physical and chemical parameters and cell count values of the samples. | ||||||||
| Parameter | Nagyborvíz | Piricske | Taploca | Berzsenyi | Kiskút | Kossuth Lajos | Szent Jakab | Polányi kút |
| T (°C) | 14.1 | 8.5 | 17.7 | 17.2 | 15 | 17.1 | 19.7 | 18.5 |
| pH | 6.18 | 6.74 | 6.08 | 6.23 | 7.19 | 6.66 | 7.04 | 6.23 |
| conductivity (μS/cm) | 2785 | 163 | 1490 | 1627 | 913 | 1185 | 581 | 1526 |
| ORP1 (mV) | 48.1 | 14.8 | 27.8 | 55.9 | 25.9 | 49.6 | 18.6 | 50.1 |
| Oxygen potential (mgl-1) | 0.63 | 5.16 | 0.39 | 0.58 | 5.25 | 4.01 | 7.07 | 0.70 |
| TOC2 (mgl-1) | 1.72 | 2.81 | 0.85 | 0.50 | 1.40 | 1 | 13 | 8.40 |
| NO3- (mgl-1) | < 0.50 | 3.10 | < 0.50 | 1.70 | 22 | 1.50 | 19 | 1.60 |
| NO2- (mgl-1) | < 0.10 | < 0.10 | < 0.10 | < 0.10 | < 0.10 | < 0.10 | < 0.10 | < 0.10 |
| NH4+ (mgl-1) | 1.38 | 0.06 | 2.66 | 0.10 | < 0.10 | 0.10 | < 0.10 | 0.50 |
| SO42- (mgl-1) | 3.9 | 6 | 2.7 | 275 | 18 | 73 | 18 | 210 |
| Fe (mgl-1) | 8.63 | 0.07 | 5.42 | 2 | < 0.10 | 1.80 | < 0.10 | 2.40 |
| PO43- (mgl-1) | 0.31 | 0.14 | 0.79 | 0.20 | 0.50 | < 0.10 | 0.10 | 0.20 |
| Cell count *ml-1 | 5.9* 104 |
20.7*104 | 9.0*104 | 5.1*104 | 5.9*104 | 1.5*104 | 2.3*104 | 1.7*104 |
Taploca, Szent Jakab and Kiskút were grouped together based in the abundance of both archaeal and bacterial OTUs, they had a common presence of the families Comamonadaceae, Sphingomonadaceae and Rhodobacteraceae. They were mainly chemoheterotrophs with fresh water is their primary habitat. They were deeply involved in carbon biogeochemical cycling with many species possess oligotrophy characters [76,77].
Hygienic of the existing microbial communities in the water samples
The results of taxon specific PCR reactions showed that in some cases water treatment would be essential before use. Legionella species, known to be widespread in natural aquatic systems, and also in aquifers and pipelines pump stations [78-80], were present in all the samples except Piricske water sample, however the samples did not contain the pathogenic L. pneumophila (Table 2).
| Table 2: Results of taxon-specific and multitemplate PCR reactions (positive: +, negative: -). | |||||||||
| Sample name | Stenotrophomonas maltophilia | Pseudomonas aeruginosa | Legionella spp | Legionella pneumophila | E.coli/coliform | Acinetobacter baumannii | NOB (nitrite-oxidizing bacteria) | ESBL (Extended Spectrum Beta-Lactamase) producer bacteria |
Macrolide resistance |
| Nagy-borvíz | + | - | + | - | - | - | + | + | - |
| Piricske | - | - | - | - | - | - | - | - | - |
| Taploca | + | - | + | - | - | - | + | - | - |
| Berzsenyi | - | - | + | - | - | - | - | - | - |
| Kiskút | + | - | + | - | - | - | - | - | - |
| Kossuth Lajos | + | - | + | - | - | - | - | - | - |
| Szent Jakab | + | - | + | - | - | - | - | - | - |
| Polányi kút | + | - | + | - | - | - | - | - | - |
Previous studies showed that Acinetobacter baumannii was detected in 38% of the groundwater supplies highlighting the possible significance of Acinetobacter spp. in groundwaters [81]. A. baumannii, though commonly associated with aquatic environments, is an opportunistic pathogen of humans therefore, this bacterium is not favoured in artificial waters (e.g. baths, drinking water, etc.). This bacterium was not present in any tested samples.
On the other hand, Stenotrophomonas maltophilia, which is an aerobic, Gram staining negative bacterium, was detected in all the water samples except Piricske and Berzsenyi by taxon specific PCR. This bacterium is ubiquitous in aquatic habitats and soils but considered as an emerging, multidrug-resistant, opportunistic pathogen bacterium [82].
NGS results revealed that Szent Jakab was the only sample that showed the presence of Enterobacteriaceae, precisely unclassified and Lelliottia members, this family was also detected in several contaminated groundwater samples in previous researches [83]. These results are of a high concern as this water is used for drinking without any treatment.
The area where Szent Jakab groundwater is located, was characterized with high vulnerability to human contamination due to the absence of confining layer, high hydraulic conductivity of the aquifer and shallow and intense groundwater flow [6].
Conclusion
Microbial cultivation independent methods confirmed the large variations in the microbial community structure among the low nutrient content water samples by presenting a comprehensive profile of the existing microbial community. In fact, nutrient-depleted aquatic environments are highly colonized by microorganisms which are key participants in the different biogeochemical cycles. Among them, methanogenic archaea: Methanobacteriaceae, Methanoperedenaceae. Sulfate reducers: Desulfobacterota, Nitrospirota. Iron oxidiser: Gallionellaceae. Nitrate reducers: Azospirilla. Ammonia oxidizers: Nitrosomonadaceae. Moreover, the existing microorganism were involved in many different metabolic types. Many are chemotrophic, chemoautotrophic and facultative phototrophic microorganisms. The high percentage of unknown sequences suggests further that many low nutrient content aquatic environments are still undiscovered and further studies are needed.
Declarations
Availability of data and materials
All data generated or analysed during this study are included in this published article or mentioned in the section: DNA extraction from the water samples and amplicon sequencing.
Competing interests
The authors declare that they have no competing interests.
Funding
This study was funded by ELTE Thematic Excellence Programme 2020 Supported by the National Research – Development and Innovation Office – TKP2020-IKA-05.
Authors contributions
MT: Wrote the main Manuscript Text. IM: Sampling and elaboration of the first 24 hours’ experiments. AT: Hydrogeological research on the samples. NS: Performed the taxon specific and multitemplate PCR experiments. RF: Material and methods plan. LJ: Performed the physiochemical experiments. ET: Supervision and research plan. All Authors read and approved the final manuscript.
Acknowledgment
The authors are thankful to Gergely Krett, Sára Szuróczki, Balázs Vajna and Attila Szabó and for their technical advice.
References
- Felipe-Sotelo M, Henshall-Bell ER, Evans NDM, Read D. Comparison of the chemical composition of British and Continental European bottled waters by multivariate analysis. J Food Compos Anal. 2015;39(3):33-42. doi: 10.1016/j.jfca.2014.10.014.
- Casanovas-Massana A, Blanch AR. Diversity of the heterotrophic microbial populations for distinguishing natural mineral waters. Int J Food Microbiol. 2012 Feb 1;153(1-2):38-44. doi: 10.1016/j.ijfoodmicro.2011.10.012. Epub 2011 Oct 29. PMID: 22094180.
- Máthé I, Táncsics A, György E, Pohner Z, Vladár P, Székely AJ, Márialigeti K. Investigation of mineral water springs of Miercurea Ciuc (Csíkszereda) region (Romania) with cultivation-dependent microbiological methods. Acta Microbiol Immunol Hung. 2010 Jun;57(2):109-22. doi: 10.1556/AMicr.57.2010.2.4. PMID: 20587384.
- Kis BM, Baciu C. The mineral waters from the Eastern Carpathians: A chemical review. Journal of Environmental Research and Protection. 2014;59.
- KL, Budai Tamás, Császár Géza, Csillag Gábor, Dudko Antonyina, Majoros G. A balaton-felvidék földtana. Angew Chemie Int Ed. 1999;6(11):951-952.
- Tóth A, Mádl-Szőnyi J. Scale-dependent evaluation of an unconfined carbonate system- practical application, consequences and significance. 2015. p.199-213.
- Rodger BB, Andrew DE, Eugene WR, Laura Bridgewater. American Public Health Association, American Water Works Association, Water Environment Federation. Standard methods for the examination of water and wastewater. 2017.
- Zsuzsa K, Judit M, Katalin B, Balázs V, Márton P, Károly M, Erika T. Critical point analysis and biocide treatment in a microbiologically contaminated water purification system of a power plant. SN Applied Sciences. 2019;1(8):1-12. doi: 10.1007/s42452-019-0740-9.
- Abbaszade G, Szabó A, Vajna B, Farkas R, Szabó C, Tóth E. Whole genome sequence analysis of Cupriavidus campinensis S14E4C, a heavy metal resistant bacterium. Mol Biol Rep. 2020 May;47(5):3973-3985. doi: 10.1007/s11033-020-05490-8. Epub 2020 May 13. PMID: 32406019; PMCID: PMC7239810.
- Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011 Oct;5(10):1571-9. doi: 10.1038/ismej.2011.41. Epub 2011 Apr 7. PMID: 21472016; PMCID: PMC3176514.
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013 Jan 7;41(1):e1. doi: 10.1093/nar/gks808. Epub 2012 Aug 28. PMID: 22933715; PMCID: PMC3592464.
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009 Dec;75(23):7537-41. doi: 10.1128/AEM.01541-09. Epub 2009 Oct 2. PMID: 19801464; PMCID: PMC2786419.
- Klára B, János B, István M, Gyöngyvér M, Tamás F, Attila S, Csaba F, Csilla A, Robert W B, Oswald JS, Adalbert B. Linking intraspecific variation in plant chemical defence with arthropod and soil bacterial community structure and N allocation. Plant Soil. 2019;444(1-2):383-397. doi: 10.1007/s11104-019-04284-7.
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011 Aug 15;27(16):2194-200. doi: 10.1093/bioinformatics/btr381. Epub 2011 Jun 23. PMID: 21700674; PMCID: PMC3150044.
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010 Jan;12(1):118-23. doi: 10.1111/j.1462-2920.2009.02051.x. Epub 2009 Aug 27. PMID: 19725865.
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013 Jan;41(Database issue):D590-6. doi: 10.1093/nar/gks1219. Epub 2012 Nov 28. PMID: 23193283; PMCID: PMC3531112.
- Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010 Jan;60(Pt 1):249-266. doi: 10.1099/ijs.0.016949-0. Epub 2009 Aug 21. PMID: 19700448.
- Bej AK, Steffan RJ, DiCesare J, Haff L, Atlas RM. Detection of coliform bacteria in water by polymerase chain reaction and gene probes. Appl Environ Microbiol. 1990;56(2):307-314. doi: 10.1128/aem.56.2.307-314.1990.
- Huang LY, Chen TL, Lu PL, Tsai CA, Cho WL, Chang FY, Fung CP, Siu LK. Dissemination of multidrug-resistant, class 1 integron-carrying Acinetobacter baumannii isolates in Taiwan. Clin Microbiol Infect. 2008 Nov;14(11):1010-9. doi: 10.1111/j.1469-0691.2008.02077.x. Erratum in: Clin Microbiol Infect. 2009 Mar;15(3):296. PMID: 19040472.
- Cloud JL, Carroll KC, Pixton P, Erali M, Hillyard DR. Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J Clin Microbiol. 2000 May;38(5):1709-12. doi: 10.1128/JCM.38.5.1709-1712.2000. PMID: 10790085; PMCID: PMC86568.
- Gallo SW, Ramos PL, Ferreira CA, Oliveira SD. A specific polymerase chain reaction method to identify Stenotrophomonas maltophilia. Mem Inst Oswaldo Cruz. 2013 May;108(3):390–1. doi: 10.1590/S0074-02762013000300020. PMID: 23778655; PMCID: PMC4005581.
- Fiume L, Bucci Sabattini MA, Poda G. Detection of Legionella pneumophila in water samples by species-specific real-time and nested PCR assays. Lett Appl Microbiol. 2005;41(6):470-5. doi: 10.1111/j.1472-765X.2005.01779.x. Erratum in: Lett Appl Microbiol. 2006 Mar;42(3):304. Bucca Sabattini, MA [corrected to Bucci Sabattini, MA]. PMID: 16305672.
- Lavenir R, Jocktane D, Laurent F, Nazaret S, Cournoyer B. Improved reliability of Pseudomonas aeruginosa PCR detection by the use of the species-specific ecfX gene target. J Microbiol Methods. 2007;70(1):20-29. doi: 10.1016/j.mimet.2007.03.008.
- Trung NT, Hien TT, Huyen TT, Quyen DT, Binh MT, Hoan PQ, Meyer CG, Velavan TP, Song le H. Simple multiplex PCR assays to detect common pathogens and associated genes encoding for acquired extended spectrum betalactamases (ESBL) or carbapenemases from surgical site specimens in Vietnam. Ann Clin Microbiol Antimicrob. 2015 Apr 12;14:23. doi: 10.1186/s12941-015-0079-z. PMID: 25890291; PMCID: PMC4399146.
- Zmantar T, Kouidhi B, Miladi H, Bakhrouf A. Detection of macrolide and disinfectant resistance genes in clinical Staphylococcus aureus and coagulase-negative staphylococci. BMC Res Notes. 2011 Oct 27;4:453. doi: 10.1186/1756-0500-4-453. PMID: 22032892; PMCID: PMC3212975.
- Albertsen M, Karst SM, Ziegler AS, Kirkegaard RH, Nielsen PH. Back to Basics--The Influence of DNA Extraction and Primer Choice on Phylogenetic Analysis of Activated Sludge Communities. PLoS One. 2015 Jul 16;10(7):e0132783. doi: 10.1371/journal.pone.0132783. PMID: 26182345; PMCID: PMC4504704.
- Oksanen AJ. Vegan. Encyclopedia of Food and Agricultural Ethics. In: Kaplan DM, editor. 2019. p.2395-2396. doi: 10.1007/978-94-024-1179-9_301576.
- Lemos LN, Fulthorpe RR, Triplett EW, Roesch LF. Rethinking microbial diversity analysis in the high throughput sequencing era. J Microbiol Methods. 2011 Jul;86(1):42-51. doi: 10.1016/j.mimet.2011.03.014. Epub 2011 Mar 30. PMID: 21457733.
- Kuznetsov SI, Dubinina GA, Lapteva NA. Biology of oligotrophic bacteria. Annu Rev Microbiol. 1979;33:377-87. doi: 10.1146/annurev.mi.33.100179.002113. PMID: 386927.
- Ho A, Di Lonardo DP, Bodelier PL. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol Ecol. 2017 Mar 1;93(3). doi: 10.1093/femsec/fix006. PMID: 28115400.
- Lopez-Fernandez M, Åström M, Bertilsson S, Dopson M. Depth and Dissolved Organic Carbon Shape Microbial Communities in Surface Influenced but Not Ancient Saline Terrestrial Aquifers. Front Microbiol. 2018 Nov 27;9:2880. doi: 10.3389/fmicb.2018.02880. PMID: 30538690; PMCID: PMC6277548.
- Gayner NJ. River bank inducement influence on a shallow groundwater microbial community and its effects on aquifer reactivity. 2018.
- Marwene T, Gorkhmaz A, Yousra S, Rózsa F, Éva Á, Laura J, Erika T. Cultivation and molecular studies to reveal the microbial communities of groundwaters discharge located in Hungary. Water. 2021;13:11. doi: 10.3390/w13111533.
- Liu X, Li M, Castelle CJ, Probst AJ, Zhou Z, Pan J, Liu Y, Banfield JF, Gu Ji D. Insights into the ecology, evolution, and metabolism of the widespread woesearchaeotal lineages. Microbiome. 2018;6(1):1-16. doi: 10.1186/s40168-018-0488-2.
- Lau MCY, Kieft TL, Kuloyo O, Linage-Alvarez B, van Heerden E, Lindsay MR, Magnabosco C, Wang W, Wiggins JB, Guo L, Perlman DH, Kyin S, Shwe HH, Harris RL, Oh Y, Yi MJ, Purtschert R, Slater GF, Wei S, Li L, Sherwood LB, Onstott TC, Ono S. An oligotrophic deep-subsurface community dependent on syntrophy is dominated by sulfur-driven autotrophic denitrifiers. Proc Natl Acad Sci. 2016;113(49):E7927-E7936. doi: 10.1073/pnas.1612244113.
- Castelle CJ, Brown CT, Anantharaman K, Probst AJ, Huang RH, Banfield JF. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat Rev Microbiol. 2018 Oct;16(10):629-645. doi: 10.1038/s41579-018-0076-2. PMID: 30181663.
- Hu W, Pan J, Wang B, Guo J, Li M, Xu M. Metagenomic insights into the metabolism and evolution of a new Thermoplasmata order (Candidatus Gimiplasmatales). Environ Microbiol. 2021 Jul;23(7):3695-3709. doi: 10.1111/1462-2920.15349. Epub 2020 Dec 15. PMID: 33295091.
- Zhou Z, Liu Y, Lloyd KG, Pan J, Yang Y, Gu JD, Li M. Genomic and transcriptomic insights into the ecology and metabolism of benthic archaeal cosmopolitan, Thermoprofundales (MBG-D archaea). ISME J. 2019 Apr;13(4):885-901. doi: 10.1038/s41396-018-0321-8. Epub 2018 Dec 4. PMID: 30514872; PMCID: PMC6461988.
- Hartmann M, Grob C, Tarran GA, Martin AP, Burkill PH, Scanlan DJ, Zubkov MV. Mixotrophic basis of Atlantic oligotrophic ecosystems. Proc Natl Acad Sci U S A. 2012 Apr 10;109(15):5756-60. doi: 10.1073/pnas.1118179109. Epub 2012 Mar 26. PMID: 22451938; PMCID: PMC3326507.
- Shuming MCJ, Jinhui L, Bin L, Ran Y, Shiqing N, Zufan Z, Jianping L, Qiong J, Bing Y. Impacts of Crenarchaeota and halobacterota on sulfate reduction in the subtropical mangrove ecosystem as revealed by smdb analysis. bioRxiv. 2020;34(5):155-163. doi: 10.1101/2020.08.16.252635.
- Rios-Del Toro EE, Valenzuela EI, Ramírez JE, López-Lozano NE, Cervantes FJ. Anaerobic ammonium oxidation linked to microbial reduction of natural organic matter in marine sediments. Environ Sci Technol Lett. 2018;5(9):571-577. doi: 10.1021/acs.estlett.8b00330.
- Kerou M, Schleper C. Nitrososphaeraceae. Wiley Online Library. 2016. p.1-2. doi: 10.1002/9781118960608.fbm00265.
- Prosser JI, Nicol GW. Candidatus Nitrosotaleaceae. Bergey’s manual of systematics of archaea and bacteria. 2016;1-1. doi: 10.1002/9781118960608.fbm00264.
- Isobe K, Ikutani J, Fang YT, Yoh M, Mo JM, Suwa Y, Yoshida M, Senoo K, Otsuka S, Koba K. Highly abundant acidophilic ammonia-oxidizing archaea causes high rates of nitrification and nitrate leaching in nitrogen-saturated forest soils. Soil Biol Biochem. 2018;122:220-227. doi: 10.1016/j.soilbio.2018.04.021.
- Rosenberg E. The prokaryotes: Alphaproteobacteria and betaproteobacteria. 2013;1-1012. doi: 10.1007/978-3-642-30197-1.
- Chen M, Conroy JL, Sanford RA, Chee-Sanford JC, Connor LM, Interpreting lacustrine bulk sediment δ15N values using metagenomics in a tropical hypersaline lake system. J Paleolimnol. 2021;65(1):151-168. doi: 10.1007/s10933-020-00157-7.
- Herrmann M, Wegner CE, Taubert M, Geesink P, Lehmann K, Yan L, Lehmann R, Totsche KU, Küsel K. Predominance of Cand. Patescibacteria in Groundwater Is Caused by Their Preferential Mobilization From Soils and Flourishing Under Oligotrophic Conditions. Front Microbiol. 2019 Jun 20;10:1407. doi: 10.3389/fmicb.2019.01407. PMID: 31281301; PMCID: PMC6596338.
- Miyoshi T, Iwatsuki T, Naganuma T. Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters. Appl Environ Microbiol. 2005 Feb;71(2):1084-8. doi: 10.1128/AEM.71.2.1084-1088.2005. PMID: 15691970; PMCID: PMC546738.
- Luef B, Frischkorn KR, Wrighton KC, Holman HY, Birarda G, Thomas BC, Singh A, Williams KH, Siegerist CE, Tringe SG, Downing KH, Comolli LR, Banfield JF. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun. 2015 Feb 27;6:6372. doi: 10.1038/ncomms7372. PMID: 25721682.
- Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF, Carlson CA, Smith RD, Giovanonni SJ. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2009 Jan;3(1):93-105. doi: 10.1038/ismej.2008.83. Epub 2008 Sep 4. PMID: 18769456.
- Giovannoni SJ, Cameron Thrash J, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014 Aug;8(8):1553-65. doi: 10.1038/ismej.2014.60. Epub 2014 Apr 17. PMID: 24739623; PMCID: PMC4817614.
- Alejandro RG, Julia K N, Maliheh M, Moritz B, Frederik S, Tanja W, Sarahi LG. A genomic perspective on genome size distribution across Earth’s microbiomes reveals a tendency to gene loss. bioRxiv. 2021;1-25. doi: 10.1101/2021.01.18.427069.
- Krzmarzick MJ, Crary BB, Harding JJ, Oyerinde OO, Leri AC, Myneni SC, Novak PJ. Natural niche for organohalide-respiring Chloroflexi. Appl Environ Microbiol. 2012 Jan;78(2):393-401. doi: 10.1128/AEM.06510-11. Epub 2011 Nov 18. PMID: 22101035; PMCID: PMC3255752.
- Probst AJ, Weinmaier T, Raymann K, Perras A, Emerson JB, Rattei T, Wanner G, Klingl A, Berg IA, Yoshinaga M, Viehweger B, Hinrichs KU, Thomas BC, Meck S, Auerbach AK, Heise M, Schintlmeister A, Schmid M, Wagner M, Gribaldo S, Banfield JF, Moissl-Eichinger C. Biology of a widespread uncultivated archaeon that contributes to carbon fixation in the subsurface. Nat Commun. 2014 Nov 26;5:5497. doi: 10.1038/ncomms6497. PMID: 25425419.
- Bräuer S, Cadillo-Quiroz H, Kyrpides N, Woyke T, Goodwin L, Detter C, Podell S, Yavitt JB, Zinder SH. Genome of Methanoregula boonei 6A8 reveals adaptations to oligotrophic peatland environments. Microbiology (Reading). 2015 Aug;161(8):1572-1581. doi: 10.1099/mic.0.000117. Epub 2015 May 21. PMID: 25998264.
- Jackson BE, Bhupathiraju VK, Tanner RS, Woese CR, McInerney MJ. Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch Microbiol. 1999 Jan;171(2):107-14. doi: 10.1007/s002030050685. PMID: 9914307.
- Simon J, Kroneck PM. Microbial sulfite respiration. Adv Microb Physiol. 2013;62:45-117. doi: 10.1016/B978-0-12-410515-7.00002-0. PMID: 23481335.
- Umezawa K, Kojima H, Kato Y, Fukui M. Disproportionation of inorganic sulfur compounds by a novel autotrophic bacterium belonging to Nitrospirota. Syst Appl Microbiol. 2020 Sep;43(5):126110. doi: 10.1016/j.syapm.2020.126110. Epub 2020 Jul 2. PMID: 32847785.
- Frank YA, Kadnikov VV, Lukina AP, Banks D, Beletsky AV, Mardanov AV, Sen'kina EI, Avakyan MR, Karnachuk OV, Ravin NV. Characterization and Genome Analysis of the First Facultatively Alkaliphilic Thermodesulfovibrio Isolated from the Deep Terrestrial Subsurface. Front Microbiol. 2016 Dec 19;7:2000. doi: 10.3389/fmicb.2016.02000. PMID: 28066337; PMCID: PMC5165239.
- Okon Y. Azospirillum as a potential inoculant for agriculture. Trends Biotechnol. 1985;3(9):223-228. doi: 10.1016/0167-7799(85)90012-5.
- Saito A, Mitsui H, Hattori R, Minamisawa K, Hattori T. Slow-growing and oligotrophic soil bacteria phylogenetically close to bradyrhizobium japonicum. FEMS Microbiol Ecol. 1998;25(3):277-286. doi: 10.1016/S0168-6496(98)00009-9.
- Chrzanowski TH, Kyle M, Elser JJ, Sterner R. Element ratios and growth dynamics of bacteria in an oligotrophic Canadian shield lake. Aquat Microb Ecol. 1996;11(2):119-125. doi: 10.3354/ame011119.
- Wang J, Wang C, Li J, Bai P, Li Q, Shen M, Li R, Li T, Zhao J. Comparative Genomics of Degradative Novosphingobium Strains With Special Reference to Microcystin-Degrading Novosphingobium sp. THN1. Front Microbiol. 2018 Sep 25;9:2238. doi: 10.3389/fmicb.2018.02238. PMID: 30319567; PMCID: PMC6167471.
- White DC, Sutton SD, Ringelberg DB. The genus Sphingomonas: physiology and ecology. Curr Opin Biotechnol. 1996 Jun;7(3):301-6. doi: 10.1016/s0958-1669(96)80034-6. PMID: 8785434.
- Verma H, Dhingra GG, Sharma M, Gupta V, Negi RK, Singh Y, Lal R. Comparative genomics of Sphingopyxis spp. unravelled functional attributes. Genomics. 2020 Mar;112(2):1956-1969. doi: 10.1016/j.ygeno.2019.11.008. Epub 2019 Nov 15. PMID: 31740292.
- Park M, Jung I, Song J, Cho JC. Sphingorhabdus lacus sp. nov. and Sphingorhabdus profundilacus sp. nov., isolated from freshwater environments. Int J Syst Evol Microbiol. 2020 May;70(5):3202-3209. doi: 10.1099/ijsem.0.004155. PMID: 32320377.
- Emerson D, Field EK, Chertkov O, Davenport KW, Goodwin L, Munk C, Nolan M, Woyke T. Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol. 2013 Sep 12;4:254. doi: 10.3389/fmicb.2013.00254. PMID: 24062729; PMCID: PMC3770913.
- Atashgahi S, Aydin R, Dimitrov MR, Sipkema D, Hamonts K, Lahti L, Maphosa F, Kruse T, Saccenti E, Springael D, Dejonghe W, Smidt H. Impact of a wastewater treatment plant on microbial community composition and function in a hyporheic zone of a eutrophic river. Sci Rep. 2015 Nov 26;5:17284. doi: 10.1038/srep17284. PMID: 26607034; PMCID: PMC4660315.
- Humphreys WF. Hydrogeology and groundwater ecology: Does each inform the other? Hydrogeol J. 2009;17(1):5-21. doi: 10.1007/s10040-008-0349-3.
- Edberg SC, LeClerc H, Robertson J. Natural protection of spring and well drinking water against surface microbial contamination. II. Indicators and monitoring parameters for parasites. Crit Rev Microbiol. 1997;23(2):179-206. doi: 10.3109/10408419709115135. PMID: 9226113.
- Bekins B. Preface-groundwater and microbial processes. Hydrogeol J. 2000;8(1):2-3. doi: 10.1007/s100400050002.
- Fazi S, Amalfitano S, Venturi S, Pacini N, Vazquez E, Olaka LA, Tassi F, Crognale S, Herzsprung P, Lechtenfeld OJ, Cabassi J, Capecchiacci F, Rossetti S, Yakimov MM, Vaselli O, Harper DM, Butturini A. High concentrations of dissolved biogenic methane associated with cyanobacterial blooms in East African lake surface water. Commun Biol. 2021 Jul 7;4(1):845. doi: 10.1038/s42003-021-02365-x. PMID: 34234272; PMCID: PMC8263762.
- Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013 Aug 29;500(7464):567-70. doi: 10.1038/nature12375. Epub 2013 Jul 28. Erratum in: Nature. 2013 Sep 26;501(7468):578. PMID: 23892779.
- Timmers PH, Suarez-Zuluaga DA, van Rossem M, Diender M, Stams AJ, Plugge CM. Anaerobic oxidation of methane associated with sulfate reduction in a natural freshwater gas source. ISME J. 2016 Jun;10(6):1400-12. doi: 10.1038/ismej.2015.213. Epub 2015 Dec 4. PMID: 26636551; PMCID: PMC5029187.
- Gülay A, Çekiç Y, Musovic S, Albrechtsen HJ, Smets BF. Diversity of Iron Oxidizers in Groundwater-Fed Rapid Sand Filters: Evidence of Fe(II)-Dependent Growth by Curvibacter and Undibacterium spp. Front Microbiol. 2018 Dec 3;9:2808. doi: 10.3389/fmicb.2018.02808. PMID: 30559723; PMCID: PMC6287000.
- Vaz-Moreira I, Nunes OC, Manaia CM. Diversity and antibiotic resistance patterns of Sphingomonadaceae isolates from drinking water. Appl Environ Microbiol. 2011 Aug 15;77(16):5697-706. doi: 10.1128/AEM.00579-11. Epub 2011 Jun 24. PMID: 21705522; PMCID: PMC3165245.
- Willems A, Gillis M. Comamonadaceae. Bergey’s Man Syst Archaea Bact. 2015;1-6. doi: 10.1002/9781118960608.fbm00182.
- Devos L, Boon N, Verstraete W. Legionella pneumophila in the environment: The occurrence of a fastidious bacterium in oligotrophic conditions. Rev Environ Sci Biotechnol. 2005;4(1/2):61-74. doi: 10.1007/s11157-004-8174-1.
- Devos L, Clymans K, Boon N, Verstraete W. Evaluation of nested PCR assays for the detection of Legionella pneumophila in a wide range of aquatic samples. J Appl Microbiol. 2005;99(4):916-25. doi: 10.1111/j.1365-2672.2005.02668.x. PMID: 16162244.
- Felföldi T, Heéger Z, Vargha M, Márialigeti K. Detection of potentially pathogenic bacteria in the drinking water distribution system of a hospital in Hungary. Clin Microbiol Infect. 2010 Jan;16(1):89-92. doi: 10.1111/j.1469-0691.2009.02795.x. PMID: 19519854.
- Bifulco JM, Shirey JJ, Bissonnette GK. Detection of Acinetobacter spp. in rural drinking water supplies. Appl Environ Microbiol. 1989 Sep;55(9):2214-9. doi: 10.1128/aem.55.9.2214-2219.1989. PMID: 2529816; PMCID: PMC203058.
- Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. 2012 Jan;25(1):2-41. doi: 10.1128/CMR.00019-11. PMID: 22232370; PMCID: PMC3255966.
- Chakraborty S, Kumar RN. Assessment of groundwater quality at a MSW landfill site using standard and AHP based water quality index: a case study from Ranchi, Jharkhand, India. Environ Monit Assess. 2016 Jun;188(6):335. doi: 10.1007/s10661-016-5336-x. Epub 2016 May 7. PMID: 27155859.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.