Medicine Group . 2022 November 08;3(11):1293-1297. doi: 10.37871/jbres1595.
Prolonged Viral Shedding and Multiple Admissions Due to SARS-CoV-2-Associated Pneumonia in a Patient with Chronic Lymphocytic Leukemia: A Case Report
Sergio Espana-Cueto1#, Jose Ramon Santos1#*, Oscar Blanch-Lombarte2, Joan-Ramon Grifols3, Gemma Llados1, Cristina Lopez1, Daniel Molina-Morant1, Maria Nevot2, Edwards Pradenas2, Benjamin Trinite2, Esther Jimenez-Moyano2, Ruth Pena2, Silvia Marfil2, Mariona Parera2, Francesc Catala2, Agueda Hernandez-Rodriguez4, Antoni Escalas4,9, Ignacio Blanco5, Veronica Saludes4,6, Elisa Martro4,6, Carla Rovirosa2, Eva Riveira-Munoz2, Teresa Puig2, Christelle Ferra7, Carmen Fernandez-Arias8, Julia Blanco2,9-11, Ester Ballana2,9, Bonaventura Clotet1,2,10,11, Pere J Cardona4,9,11,13, Marc Noguera-Julian2,11, Julia Garcia-Prado2,9,11, Lourdes Mateu1,13, Marta Massanella2,11 and Roger Paredes1,2,11,14
- SARS-CoV-2
- Prolonged viral shedding
- Chronic lymphocytic leukemia
Abstract
We describe a case of a patient who experienced recurrent COVID-19 pneumonia over a period of 123 days. Neither remdesivir nor convalescent plasma were temporally associated with viral clearance or increased plasma neutralization capacity. Antibody levels remained low until day 151 and cellular immunity increased overtime with no viral evolution.
Abbreviations
PaO2/FiO2 ratio: Ratio of arterial oxygen partial pressure (PaO2 in mmHg) to fractional inspired oxygen (FiO2 expressed as a fraction, not a percentage); CRP: C-Reactive Protein; LDH: Lactate Dehydrogenase; IL-6: Interleukin 6; DXM: Dexamethasone; CP: Convalescent Plasma; VL: Viral Load
Introduction
While most non-immunocompromised subjects effectively clear SARS-CoV-2 infection in less than 10 days [1], there are several reports of prolonged viral shedding associated with immunosuppressive status [2,3]. The adaptive immune system (B cells, CD4+ and CD8+ T-cells) and neutralizing antibodies are important factors to control viral infections, including SARS-CoV-2 [4]. Chronic Lymphocytic Leukemia (CLL) is a cause of immunosuppressive status due to impaired humoral and cellular immunity [5], involving both innate and adaptive immune responses. In addition, type, duration, number and combinations of therapies, including rituximab or venetoclax [6,7], also contribute to immune suppression in subjects with CLL.
Many uncertainties persist around the management of COVID-19 in immunocompromised patients with hematological malignancies [8]. We report a case of recurrent SARS-CoV-2 infection in a patient with CLL with previous exposure to rituximab and following third-line treatment with venetoclax, a selective BCL-2 inhibitor molecule and BH-3-only mimetic [6]. We describe the clinical characteristics of this patient, as well as the evolution of humoral and cellular mediated immune responses over time.
Material and Methods
SARS-CoV-2 viral load was quantified from nasopharyngeal swab samples by reverse transcription quantitative PCR (RT-qPCR). The presence of anti-SARS-CoV-2 antibodies in plasma samples was evaluated using an in-house sandwich-ELISA. Both HIV reporter pseudoviruses expressing SARS-CoV-2 spike protein and luciferase were used for the virus neutralization assay. The T-cell response was determined by IFNγ ELISpot assay using 30 peptide pools covering the spike, membrane, nucleocapsid and nsp3 regions of SARS-CoV-2 proteome. Viral genome sequencing was performed to evaluate the development of spike mutations and intra-host virus evolution (supplementary methods). Every convalescent plasma unit was obtained by plasmapheresis procedure from donors, containing a volume of 220-315 mL, anti-SARS-CoV-2 anti-Spike IgG (≥ 20,000 AU/ml) and residual leukocytes (< 1 x 10E9/L). Blood group and Rh (D) and negative infectious markers were identified prior to infusion, according to current regulations and recommendations. The patient provided written informed consent before sampling.
Case Description
Patient was a 64-year-old female with medical history of chronic hypertension and left lung tumorectomy secondary to stage Ib lung adenocarcinoma, in remission since 2015. She was diagnosed with CLL in 2015 and was treated with rituximab, fludarabine and cyclophosphamide in 2016, acalabrutinib between September 2017 and March 2020, and venetoclax (400 mg QD) from July 2020 onwards.
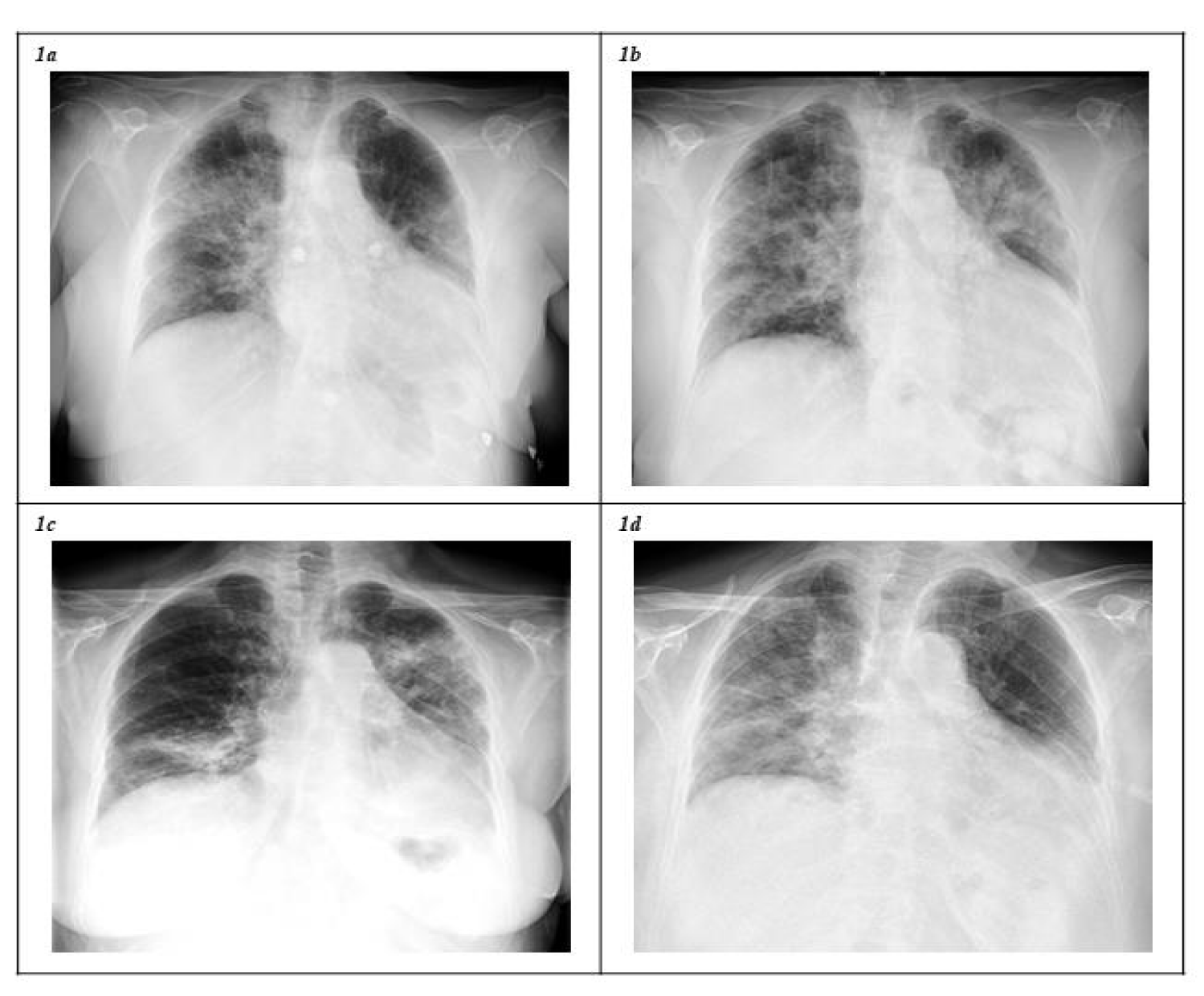
A nasopharyngeal RT-PCR test for SARS-CoV-2 was positive on February 7 2021, following a contact-tracing study. The patient started presenting with fever up to 39ºC and cough on February 18. After experiencing worsening symptoms, she was admitted on February 27 with bilateral opacities revealed by chest X-ray (Figure 1a). Treatment with remdesivir (five days), dexamethasone (6 mg/day) and prophylactic enoxaparin were administered. Subsequently, the patient experienced clinical improvement and was discharged on March 3, her treatment thereafter being dexamethasone for ten days and prophylactic enoxaparin.
Forty-eight hours after discharge, the patient started feeling shortness of breath on exertion, asthenia and fever up to 39.3ºC, being admitted with ARDS and new radiographic opacities (Figure 1b) on March 14. RT-PCR testing for SARS-CoV-2 was still positive (Cycle Threshold (Ct): 22). During the following days, non-invasive mechanical ventilation was started with a fraction of inspired oxygen (FiO2) of 100%. Oxygen, remdesivir and dexamethasone were started again, and prophylactic enoxaparin was maintained. The subsequent evolution was satisfactory, and she was discharged for a second time on March 20, with radiological improvement and without supplementary oxygen.
On April 1, the patient again experienced fever up to 39ºC, chills, profuse sweating, asthenia and cough. She was admitted to hospital for a third time on April 7 because of a new COVID-19 pneumonia and ARDS (Figure 1c). A SARS-CoV-2 RT-PCR test was positive (Ct: 23.5). The same treatment regimen with oxygen, remdesivir and dexamethasone was followed and, additionally, one unit of Convalescent Plasma (CP) was administered on April 10. After experiencing progressive improvement, she was discharged asymptomatic on April 18.
Once again, on April 27 she presented with asthenia, dysthermia, dyspnea and cough. A new RT-PCR test result was positive (Ct: 25.7) and, due to clinical aggravation, she was admitted on April 30 with ARDS and radiological progression compared to previous findings (Figure 1d). Supplementary oxygen, remdesivir, dexamethasone and prophylaxis with enoxaparin were administered, followed by a second unit of CP on April 29. She was discharged on May 5 after a favorable evolution, with two more scheduled CP infusions every fourteen days. Finally, on July 6 2021, 151 days after the first positive RT-PCR test for SARS-CoV-2 and following four hospital stays in which the patient underwent the standard treatment regimen for COVID-19 pneumonia and the administration of four units of CP, a non-detectable RNA result was confirmed, accompanied by complete clinical recovery.
Discussion
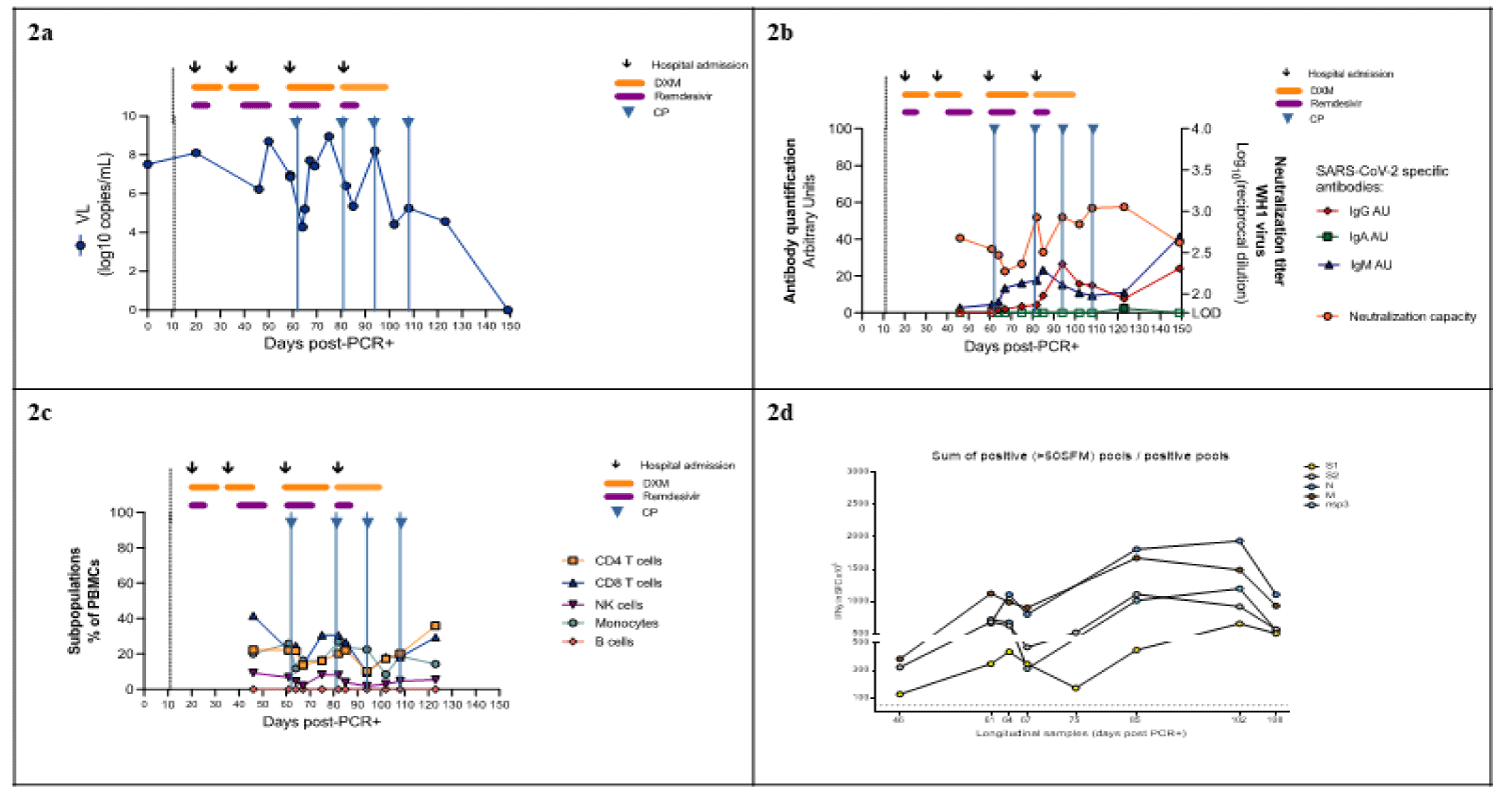
In the case of SARS-CoV-2 infection described here, whereas consecutive treatments with remdesivir and dexamethasone in accordance with international guidelines were associated with clear clinical improvements consistent with clinical trial results [9,10], neither remdesivir nor CP infusion were associated with SARS-CoV-2 Viral Load (VL) reductions (Figure 2a). Nasopharyngeal RT-PCRs showed fluctuating but persistent, high-level viremia up to 123 days after the initial diagnosis, becoming aviremic by day 151. In fact, SARS-CoV-2 VL at hospital discharge was higher than VL at admission in at least three of the four COVID-19 episodes. This incomplete viral clearance of SARS-CoV-2 could be due to secondary impairments in the humoral and cellular adaptative immune responses associated with CLL, as well as previous treatments including rituximab [7]. In addition, immunosuppressive effects of dexamethasone could have also contributed to increase and/or prolong viral shedding [11].
Limited data from case reports and observational studies suggest a potential benefit of CP in immunosuppressed patients, including those with CLL [6,12]. In our patient, however, IgG/A/M anti-S2 + RBD SARS-CoV-2 antibodies (in-house sandwich ELISA) remained low until day 151, when IgG and IgM (but not IgA) increased. CP infusion was not associated with increased plasma neutralization capacity as titers remained mostly < 3.0log10 (Figure 2b), and it did not have a discernible clinical impact, which calls into question its usefulness for preventing new relapses. This is consistent with results from clinical trials in which CP was not effective for the treatment of hospitalized patients with COVID-19 [13].
A progressive increase in serum levels of first IgM and then IgG was found. This, however, was observed later than usually observed in non-immunosuppressed subjects [14] and in spite of there being undetectable levels of B lymphocyte in peripheral blood during monitoring. This suggests that B-lymphocytes located in compartments other than the blood, despite being dysfunctional, may be able to produce antibodies against SARS-CoV-2, albeit later than in the immunocompetent population (Figure 2c). In addition, an increase in the cell-mediated immune response both in magnitude and breadth was observed starting from day 105 after the first positive RT-PCR test. This improvement took place parallel to the increase in humoral immunity and coincided with a complete viral shedding and the patient’s clinical improvement, with a response to nucleocapsid proteins proving to be greater than the response to other viral proteins (Figure 2d).
Patients with underlying immunosuppression may constitute a scenario that facilitates viral evolution, as has been reported in several cases [15]. Moreover, intra-host evolution and the development of viruses with mutations in the spike glycoprotein have been considered to be one mechanism for the emergence of SARS-CoV-2 variants [16]. In our patient, virus genome sequencing from a sample obtained at first admission was performed, and an Alpha Variant (officially referred to as B.1.1.7) was identified. Additionally, several samples were subsequently obtained and sequenced at every admission at the hospital, revealing the presence of four mutations in four regions (ORF1a:Q3332L, ORF1a:T1001l, ORF1b:T2537l, ORF7a:Q62S) which appeared alternately without any clear evidence of viral evolution over the course of the infection. This lack of intra-host virus evolution suggests an absence of endogenous or exogenous selective pressure on the virus in upper respiratory airways in this immunosuppressed patient.
In summary, subjects with CLL are susceptible to developing a prolonged SARS-CoV-2 infection with the consequent pneumonia and ARDS, even on multiple occasions. Similarly, to what occurs in the general population, consecutive treatments with remdesivir and dexamethasone, as well as prophylaxis with enoxaparin and supplementary oxygen, seemed to be useful to treat pneumonia and ARDS episodes, while delayed and effective humoral and cellular-mediated responses led to definitive viral clearance. By contrast, infusion of CP did not seem to play a useful role in achieving good clinical outcomes. Well-designed randomized clinical trials of new antiviral drugs are urgently needed to provide personalized treatment for COVID-19 hospitalized subjects at risk of viral clearance delay, as well as to explore whether SARS-CoV-2 RNA levels could predict clinical outcomes and recurrences in immunosuppressed patients.
Funding Information
This study was supported by The Fight AIDS and Infectious Diseases FLS Foundation (Badalona, Spain), Institut de Recerca de la SIDA-IrsiCaixa (Badalona, Spain) and Fundació La Marató de TV3 (202126-30-31). In addion, it was partially funded by Grifols and the crowdfunding initiatives “https://www.yomecorono.com”, BonPreu/Esclat and Correos as well as by grants from National Health Institute Carlos III (ISCIII) COV20/00660, and RETIC RD16/0025/0041 (Co-funded by European Regional Development Fund/European Social Fund) for JGP. OBL was supported by a grant for Catalan Government and the European Social Fund AGAUR-FI_B 00582 Ph.D. fellowship.
Conflict of Interest
J. R. Santos has received research funding, consultancy fees and lecture sponsorships from and has served on advisory boards for Gilead Sciences, Janssen-Cilag, Merck Sharp & Dohme and ViiV Healthcare. R. Paredes has received research funding and consultancy fees from and has served on advisory boards for Gilead Sciences, Merck Sharp & Dohme, ViiV Healthcare, Theratechnologies Inc., and Eli Lilly and Company. E. Martró has received research funding from Gilead Sciences and lecture sponsorships from Gilead Sciences and Merck Sharp & Dohme. J Blanco has received research funding and consultancy fees from Nesapor, Hipra and Merck Sharp & Dohme, and is founder and CEO of Albajuna Therapeutics. The authors declare that they have no competing financial interests or personal relationships that could have influenced the work reported in this paper.
Acknowledgment
We are grateful to Michael Kennedy-Scanlon for proofreading assistance, and Ruth Toledo and Marta Font for their assistance in coordinating the sampling and to the technical staff of IrsiCaixa for sample processing. We thank all members of the Can Ruti Sequencing Hub for their help in generating part of the SARS-CoV-2 sequencing data. Finally, the authors thank the CERCA Programme/Generalitat de Catalunya for institutional support and the Foundation Dormeur. We would like to thank everyone who submitted sequences and metadata to GISAID.
References
- Nakajima Y, Ogai A, Furukawa K, Arai R, Anan R, Nakano Y, Kurihara Y, Shimizu H, Misaki T, Okabe N. Prolonged viral shedding of SARS-CoV-2 in an immunocompromised patient. J Infect Chemother. 2021 Feb;27(2):387-389. doi: 10.1016/j.jiac.2020.12.001. Epub 2020 Dec 4. PMID: 33328135; PMCID: PMC7836222.
- Tarhini H, Recoing A, Bridier-Nahmias A, Rahi M, Lambert C, Martres P, Lucet JC, Rioux C, Bouzid D, Lebourgeois S, Descamps D, Yazdanpanah Y, Le Hingrat Q, Lescure FX, Visseaux B. Long-Term Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infectiousness Among Three Immunocompromised Patients: From Prolonged Viral Shedding to SARS-CoV-2 Superinfection. J Infect Dis. 2021 May 20;223(9):1522-1527. doi: 10.1093/infdis/jiab075. PMID: 33556961; PMCID: PMC7928754.
- Ferrari S, Caprioli C, Weber A, Rambaldi A, Lussana F. Convalescent hyperimmune plasma for chemo-immunotherapy induced immunodeficiency in COVID-19 patients with hematological malignancies. Leuk Lymphoma. 2021 Jun;62(6):1490-1496. doi: 10.1080/10428194.2021.1872070. Epub 2021 Jan 18. PMID: 33461387; PMCID: PMC7832449.
- Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021 Feb 18;184(4):861-880. doi: 10.1016/j.cell.2021.01.007. Epub 2021 Jan 12. PMID: 33497610; PMCID: PMC7803150.
- Ginaldi L, De Martinis M, Matutes E, Farahat N, Morilla R, Catovsky D. Levels of expression of CD19 and CD20 in chronic B cell leukaemias. J Clin Pathol. 1998 May;51(5):364-9. doi: 10.1136/jcp.51.5.364. PMID: 9708202; PMCID: PMC500695.
- Juárez-Salcedo LM, Desai V, Dalia S. Venetoclax: evidence to date and clinical potential. Drugs Context. 2019 Oct 9;8:212574. doi: 10.7573/dic.212574. PMID: 31645879; PMCID: PMC6788387.
- Djaldetti M, Leibovitch C, Ganelin-Cohen E, Bessler H. Rituximab modifies peripheral blood mononuclear cells immune responses. Int J Immunol Immunother. 2019;6:037. doi: 10.23937/2378-3672/1410037.
- Martín-Moro F, Marquet J, Piris M, Michael BM, Sáez AJ, Corona M, Jiménez C, Astibia B, García I, Rodríguez E, García-Hoz C, Fortún-Abete J, Herrera P, López-Jiménez J. Survival study of hospitalised patients with concurrent COVID-19 and haematological malignancies. Br J Haematol. 2020 Jul;190(1):e16-e20. doi: 10.1111/bjh.16801. Epub 2020 May 27. PMID: 32379921; PMCID: PMC7267398.
- Beigel JH, Tomashek KM, Dodd LE. Remdesivir for the Treatment of Covid-19 - Preliminary Report. Reply. N Engl J Med. 2020 Sep 3;383(10):994. doi: 10.1056/NEJMc2022236. Epub 2020 Jul 10. PMID: 32649078.
- RECOVERY Collaborative Group, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, Elmahi E, Prudon B, Green C, Felton T, Chadwick D, Rege K, Fegan C, Chappell LC, Faust SN, Jaki T, Jeffery K, Montgomery A, Rowan K, Juszczak E, Baillie JK, Haynes R, Landray MJ. Dexamethasone in Hospitalized Patients with Covid-19. N Engl J Med. 2021 Feb 25;384(8):693-704. doi: 10.1056/NEJMoa2021436. Epub 2020 Jul 17. PMID: 32678530; PMCID: PMC7383595.
- Cogliati Dezza F, Oliva A, Cancelli F, Savelloni G, Valeri S, Mauro V, Calabretto M, Russo G, Venditti M, Turriziani O, Mastroianni CM. Determinants of prolonged viral RNA shedding in hospitalized patients with SARS-CoV-2 infection. Diagn Microbiol Infect Dis. 2021 Jun;100(2):115347. doi: 10.1016/j.diagmicrobio.2021.115347. Epub 2021 Feb 12. PMID: 33639375; PMCID: PMC7879029.
- Clark E, Guilpain P, Filip IL, Pansu N, Le Bihan C, Cartron G, Tchernonog E, Roubille C, Morquin D, Makinson A, Tuaillon E, Le Moing V. Convalescent plasma for persisting COVID-19 following therapeutic lymphocyte depletion: a report of rapid recovery. Br J Haematol. 2020 Aug;190(3):e154-e156. doi: 10.1111/bjh.16981. Epub 2020 Jul 16. PMID: 32593180; PMCID: PMC7361823.
- RECOVERY Collaborative Group. Convalescent plasma in patients admitted to hospital with COVID-19 (RECOVERY): a randomised controlled, open-label, platform trial. Lancet. 2021 May 29;397(10289):2049-2059. doi: 10.1016/S0140-6736(21)00897-7. Epub 2021 May 14. PMID: 34000257; PMCID: PMC8121538.
- Tan AT, Linster M, Tan CW, Le Bert N, Chia WN, Kunasegaran K, Zhuang Y, Tham CYL, Chia A, Smith GJD, Young B, Kalimuddin S, Low JGH, Lye D, Wang LF, Bertoletti A. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021 Feb 9;34(6):108728. doi: 10.1016/j.celrep.2021.108728. Epub 2021 Jan 21. PMID: 33516277; PMCID: PMC7826084.
- Avanzato VA, Matson MJ, Seifert SN, Pryce R, Williamson BN, Anzick SL, Barbian K, Judson SD, Fischer ER, Martens C, Bowden TA, de Wit E, Riedo FX, Munster VJ. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell. 2020 Dec 23;183(7):1901-1912.e9. doi: 10.1016/j.cell.2020.10.049. Epub 2020 Nov 4. PMID: 33248470; PMCID: PMC7640888.
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao NY, Korsman S, Davies MA, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Kosakovsky Pond SL, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021 Apr;592(7854):438-443. doi: 10.1038/s41586-021-03402-9. Epub 2021 Mar 9. PMID: 33690265.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.