Biology Group . 2022 April 30;3(4):445-452. doi: 10.37871/jbres1463.
Extra Chromosomal Circular DNA: Recent Advances in Research
Safir Ullah Khan1* and Munir Ullah Khan2
2Department of Polymer Science and Engineering, Zhejiang University, Hangzhou, China
- Extrachromosomal cyclic DNA
- Tumor
- Tumor evolution
Abstract
Extrachromosomal circular DNA (eccDNA) is a circular DNA molecule outside of eukaryotic staining, in which DNA forms in the genome or exogenous DNA in the cell. eccDNA is a special class of genetic material that can carry complete genes encoding functional proteins or RNA. Studies have shown that eccDNA can participate in various physiological and pathological processes in a special way, such as aging and the occurrence of tumors. This paper reviews the latest research progress of eccDNA and further expounds on the relationship between eccDNA and tumors.
Introduction
DNA molecules in eukaryotes have long been thought to exist primarily in a linear form in the nucleus [1]. However, in 1965, Hotta, et al. [2] found extrachromosomal circular DNA (eccDNA) in germ cells of wheat and pigs. For a century, however, eccDNA has been regarded as a piece of junk with no biological function. Until recently, several studies have found that eccDNA is ubiquitous in both normal and tumor cells and is involved in tumors' occurrence and development, especially. Compared with linear chromosomal DNA, eccDNA has an open structure and carries active protein modification, which can mediate the long-distance interaction between genes and greatly enhance the transcriptional activity of genes [3,4]. In this paper, the characteristics, research history, classification and functions of eccDNA were reviewed, and the relationship between ecDNA and tumors was deeply discussed, providing ideas for related research on eccDNA, especially the development of tumor treatment methods.
Characteristic of eccDNA
eccDNA is a circular DNA molecule that swims out of chromosomes and is ubiquitous in eukaryotes. As an extra chromatic genetic element that increases genetic heterogeneity and phenotypic difference in eukaryotes, eccDNA is widely distributed and can be detected in humans, mice, yeast, fruit flies, Arabidopsis thaliana and other organisms [5-7]. The size, abundance and sequence of eccDNA are different in different tissues, cells and individuals. In addition, the content of eccDNA is also regulated by various factors such as development and metabolism [8,9]. Recent studies have shown that eccDNA is widely present in tumor cells and can be used as a carrier for oncogene amplification. As some eccDNA does not follow Mendel's law of allocation, eccDNA carrying oncogenes and related drug resistance genes can accumulate rapidly and in large quantities in tumors, thus playing a crucial role in the occurrence and progression of tumors [10].
The size of eccDNA ranges from hundreds of bp to tens of Mb [8,11] and mainly comes from genomic or foreign DNA. eccDNA can be generated by repeated or non-repeated sequences, coding regions or non-coding regions derived from different chromosomal loci in the genome [5]. For example, Storlazz and colleagues [12] found that the deletion of MYC gene on chromosome 8 in patients with AML appeared in eccDNA, which not only indicated that the sequence of eccDNA could be derived from the genome but also suggested that eccDNA was closely related to the occurrence of Acute Myeloid Leukemia (AML). In addition, eccDNA can also be composed of chromosomal DNA and viral genome [13]. For example, eccDNA containing HCV homologous sequences can be detected from peripheral blood mononuclear cells of Hepatitis C Virus (HCV) negative patients [14]. eccDNA can also be reintegrated into the genome in the form of Homogeneous Staining Regions (HSR), which may destroy the expression of tumor suppressor genes [11,15]. eccDNA is closely related to tumors. SpcDNA, telomere rings, ecDNA and microDNA, have been found in tumors [11,16]. Among them, microDNA can be released into circulation by tumor tissues, suggesting that eccDNA may be used as a biomarker for the diagnosis and prognosis of malignant tumors [17].
Brief Research History of eccDNA
In the winter of 1868, Friedrich Miescher, a young Swiss doctor, extracted an acidic compound rich in phosphorus from white blood cells and named it "nucleoplasm," which was the first discovery of DNA [18]. Then scientists began a series of studies around DNA. STAHL believed that DNA in higher organisms might be organized into a series of circular structures, and the discovery of circular DNA in bacteria and viruses suggested the correctness of this conjecture. It was not until 1965 that Hotta, et al. [2] reported the identification of circular DNA of different sizes in wheat nuclei and pig sperm that this conjecture was officially proved. In the same year, Cox, et al. [19] reported that eccDNA was found in neuroblastoma of different sizes and quantities in different cells. Around the 1980s, with the development and maturity of molecular cloning and other technologies, scientists have successively discovered oncogenes in different samples (e.g., MYCN and EGFR can be amplified in the form of eccDNA [20], and some carcinogens and DNA replication inhibitors have also been found to promote the formation of eccDNA [5,21].
In 2012, Shibata, et al. [22] identified a small circular DNA molecule with a length of 200-400 nt, namely microDNA, in mammalian cells. In 2017, Turner, et al. [23] analyzed 17 different types of cancer cells by whole-genome sequencing, structural modeling and cytogenetic analysis. They found that eccDNA appeared in almost half of the cancer cells, and the occurrence frequency of eccDNA in different types of cancer was significantly different. In 2018, Moller, et al. [24] studied blood and muscle samples from 16 healthy people and detected about 100,000 types of eccDNA, about half of which contained complete genes or gene fragments.
Classification of eccDNA
eccDNA is classified into different types based on its source, size, and sequence characteristics, for example, MicroDNA, ecDNA (Extrachromosomal rDNA Circle (ERC), Telomeric circle, T-circle), etc… (Table 1) [5,6].
| Table 1: Characteristics of eccDNA. | ||||
| Classification of eccDNA | Size | Characteristic | Potential function | Reference |
| MicroDNA | 100-400 bp | Mostly from non-repetitive sequences | Regulate gene expression | [25] |
| ecDNA | 1-3 Mb | Accessible chromatin structure | Promote tumor deterioration | [20] |
| ERC | 19.3-40.4 Kb | Contains rDNA units | Related to aging | [8] |
| t-circle | Multiples of 738 bp | Involves in alternative lengthening of telomeres | Maintain telomere length | [6] |
ecDNA
ecDNA is available in both monomer and catamaran forms .Its liposomal form is formed by chromatin fibers joining two identical sister monomers and a microsomal [26,27]. ecDNA is ubiquitous in cancer cells and often contains proto-oncogenes and drug-resistant genes that can drive the occurrence of cancer playing a crucial role in promoting the heterogeneity and progression of tumors [20]. ecDNA is most common in neuroblastoma, esophageal cancer and squamous cell lung cancer, according to the study. Cells need to use a series of proteins such as transcription factors to conduct chromatin remodeling before recognizing and reading part of DNA genetic instructions in the nucleus [28]. ecDNA is not highly compressed. Its structure is loose and has a strong openness because this transcription factor is easy to access the cis-acting elements such as promoters.
microDNA
In identifying intramolecular homologous recombination sites, Shibata and colleagues [22] isolated a novel type of eccDNA, i.e., microDNA, from the brain cell nuclei of embryonic mice. microDNA is present in many tissues as single and double-stranded and can be released by tissues into the circulatory system. MicroDNA was 60-2 000 bp in size, and about 84% of microDNA was concentrated in 100-400 bp. By enriching circular DNA, cutting, cloning, sequencing and identification of ligand markers produced by DNA cyclization, it was found that the vast majority of microDNA came from non-repeating sequences of the gene set [22,29].
In addition, ecDNA has a loosely arranged nucleosome structure, indicating that microDNA can be generated from the genome sequence and possibly transformed from ecDNA. In other words, to meet the needs of cell survival, different types of eccDNA may be able to transform into each other and perform different regulatory functions in the face of environmental stress. Studies have found that gene-specific CpG dinucleotides on some promoters can be modified by methylation with age [30]. Paulsen, et al. [25] synthesized microDNA according to known microDNA sequences and found that these microDNA mimics could express functional microRNA and play the role of gene silencing. This suggests that microDNA transcription products may inhibit gene expression and form a functional network with other types of eccDNA, thus affecting the balance of gene quantity and function in the body.
ERC
ERC is a self-replicating circular DNA molecule that contains one or more rDNA units and lacks centromeres [31,32]. It can be produced by homologous recombination of ribosomal DNA and serve as an rRNA transcription template. Ribosomal DNA (rDNA) is the sequence of genes encoding the rRNA of ribosomal components. rDNA is highly unstable and exists as tandem repeating units [33]. Each rDNA unit has a Replication Fork Barrier (RFB) to avoid collisions between DNA replication forks and rDNA transcription complexes [34]. The stalled replication fork is very unstable, and the binding of replication fork blocking protein to RFB can induce DNA Double-Strand Break (DSB) at a high frequency [35].
If broken rDNA is repaired by intra-chromatid recombination, erCS containing one or more rDNA units can be generated from repeated rDNA sites on chromatids [36].Recent studies have suggested that ERC may be involved in cell senescence, but the relationship between ERC and senescence remains controversial. In 1997, Sinclair and Guarente [32] found that ERC accumulated in aging yeast in large quantities and inferred that ERC was a pro-aging factor. The number of ERC produced by the fOB1 mutant in yeast was reduced, and its lifespan was indeed extended [37]. The evidence supports the hypothesis that ERC promotes aging and shortens life expectancy. However, Ganley, et al. [38] observed that a decrease in ERC level would lead to a shortened life span.
ERC can maintain the stability of rDNA structure and the dynamic balance of the total number [35], and at the same time participate in the regulation of cell senescence, suggesting that the occurrence of senescence may be influenced by the genomic stability controlled by environmental factors and internal factors [39].
T cell receptor deletion ring
The precursor T cells migrated from the bone marrow into the thymus mature into primary T cells after positive and negative selection. In this process, the acquisition of functional T Cell Receptor (TCR) is the key to T cell maturation, which is crucial for T cell activation and subsequent differentiation [40]. The maturation of TCR requires V (D) J rearrangement. In this process, the V, D, and J genes assemble into the T-cell receptor V region capable of recognizing antigens [41-43]. In each tightly regulated gene rearrangement process, nucleic acid fragments can be deleted when specific gene fragments are linked to form stable eccDNA fragments, namely the so-called T-cell receptor Excision Circles (TRECs) [44].
TREC has many applications, such as evaluating thymus function after hematopoietic stem cell transplantation and detecting diseases related to T cell number and function abnormalities [45,46]. The number of TRECs is affected by many factors, such as thymus output function, initial T cell lifespan, peripheral blood T cell division and cell death, and genetic differences. Therefore, although TRECs can be used to measure thymus output, the interpretation of TRECs data in healthy people and patients still needs careful consideration [42].
eccDNA Formation Mechanisms
Currently, researchers only attempt to explain the causes of eccDNA generation by observing the sequence characteristics of eccDNA and studying the factors that promote or inhibit the formation of circular DNA [20]. However, the exact molecular mechanism of eccDNA formation is still unknown, so more work is needed. The generation of circular DNA is a complex process of multi-factor synergy [5]. In this process, DNA replication, RNA substitution, gene rearrangement and DNA Damage Repair (DDR) can all affect its formation [11,22,44]. eccDNA formation is affected by gene stability. Studies have shown that after cells are exposed to DNA damage agents such as methyl mesylate, the level of eccDNA in cells is significantly increased [47]. Mitogen-Activated Protein Kinase (MAPK) signaling pathway related to cell proliferation is often abnormally activated in malignant tumor cells. Sun, et al. [48] found that when tumor cells were treated with mapK-ERK1/2 inhibitors U0126 and PD98059 to inhibit the constitutive phosphorylation of ERK1/2, the number of DMs decreased significantly [29].
Since most eccDNA contains chromosomal DNA derived from tandem repeat sequences, homologous recombination may be involved in generating eccDNA as a common mechanism [49]. Cai, et al. [50] found that in methotrexate-resistant cells containing DMs, the attenuation of homologous recombination activity led to a significant decrease in the number of DMs. Dillon, et al. [29] found that the MSH3 DNA mismatch repair protein promotes microDNA formation through homologous recombination. In 1939, the Breakage-Fusion-Bridge (BFB) cycle (also breakage-rejoining-bridge cycle) was first proposed [51]. Recent studies suggested that the increase in gene copy number on the same chromosome caused by DNA damage during each mitosis resulted in gene amplification in the form of HSR or ecDNA [6,51,52]. This theory explains why amplified genes are often localized in DMs and HSR. Subsequently, other research groups also proposed a translocation deletion amplification model, ODIRA model and "add-body" model to explain the formation of eccDNA one after another [5,6,53].
ecDNA and Tumors
Help tumor evolution
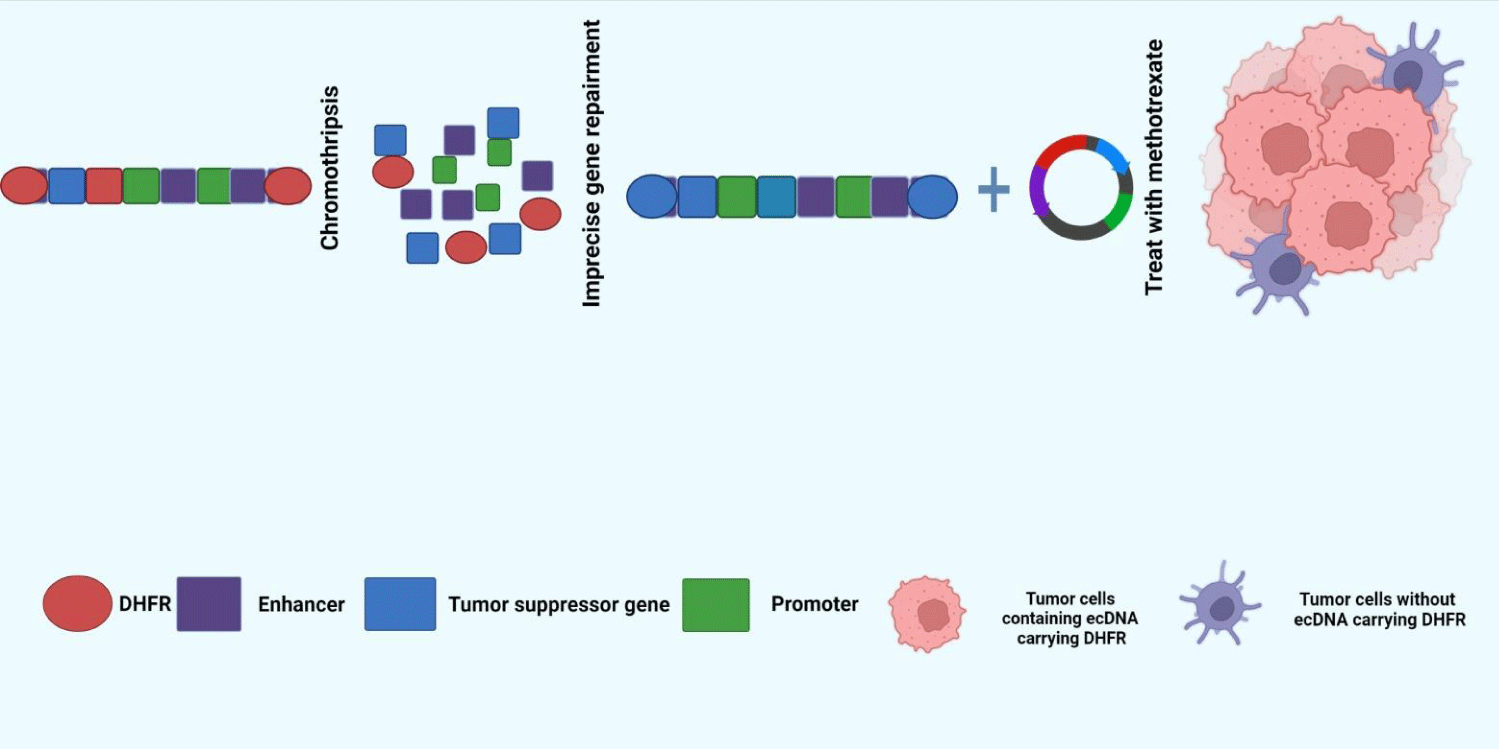
The genome is in a dynamic process of continuous change, and this instability is the driving force of spontaneous mutation of DNA accumulation in the body and the "fuel" of intracellular heterogeneity [53]. Gene mutations are influenced by exogenous and endogenous factors and can accumulate slowly over time in a progressive manner [54]. In addition, there are also big explosive mutations in cells, such as chromosome fragmentation [54]. Fragmentation is a catastrophic event of chromosomal aberration that occurs in cells and is marked by clusters of large-scale gene rearrangements. In this process, one or more chromosomes are broken in a short period, and thousands of fragments are spliced together arbitrarily to form "patch" -like chromosomes. At the same time, ecDNA may be dissociated and cycled with some genes' imprecise DNA repair mechanism [15,55].
ecDNA, as a by-product of chromosome fragmentation, mostly exists in cancer cells characterized by genetic abnormalities [23]. Oncogenes amplified by ecDNA can give survival advantages to tumor clones carrying them and drive high-level amplification of tumor clones [23]. A classic example of this is the acquisition of drug resistance by tumor cells (Figure 1). Dihydrofolate Reductase (DHFR) gene is a gene that reduces the sensitivity of cancer cells to methotrexate (MTX), often through two main topological structures: free ecDNA and HSR amplification located on chromophores. Compared with HSR, ecDNA can lead to a higher level of DHFR transcription, giving cancer cells a survival advantage and ultimately leading to adverse chemotherapy results [6].
Unlike chromosomal DNA, ecDNA lacking centromere and kinetosomes are randomly assigned to progeny cells because it is not dragged by spindle filaments [56]. This helps cancer cells accumulate large copies of a gene quickly and in a short time. If the gene is the proto-oncogene that drives cancer, then the cancer cell may, under certain selective pressures, gain a superior survival advantage and quickly develop into a dominant clone. At the same time, the randomness of ecDNA allocation makes different daughter cells carry different numbers of ecDNA containing different genes, which will lead to the simultaneous coexistence of cancer cells with different genetic characteristics, leading to the emergence of tumor heterogeneity [57]. In conclusion, ecDNA contributes to tumor survival and dynamic evolution, which is an inevitable challenge for future clinical trial design and drug development.
eccDNA and oncogene expression
For many years, when studying tumor-driven events, attention has generally focused on changes in the coding regions of identified genes [58], while chromatin topology, enhancers, and some non-coding regions beyond oncogene boundaries have not been studied [4]. Recent studies have shown that proto-oncogenes and enhancers can cyclize together to form ecDNA, which on the one hand, weakens the inhibitory effect of the repressor of gene expression and, on the other hand, strengthens the enhancer effect, thus promoting the expression of proto-oncogenes. For example, Morton, et al. [4] found that functional enhancers in glioblastoma often cyclized with oncogenes such as EGFR to weaken the inhibitory effect of suppressors, enhance the enhancing effect of enhancers, and improve the transcription efficiency of oncogenes. In addition, ecDNA-forming oncogenes tend to carry active histone modifications (such as H3K9Ac, H3K4me3, and H3K79me2) on their histone proteins, with few histone modifications that inhibit gene expression [59]. Wu, et al. [3] detected various active histone protein markers on ecDNA using high-throughput sequencing.
Morphological transformation of ecDNA
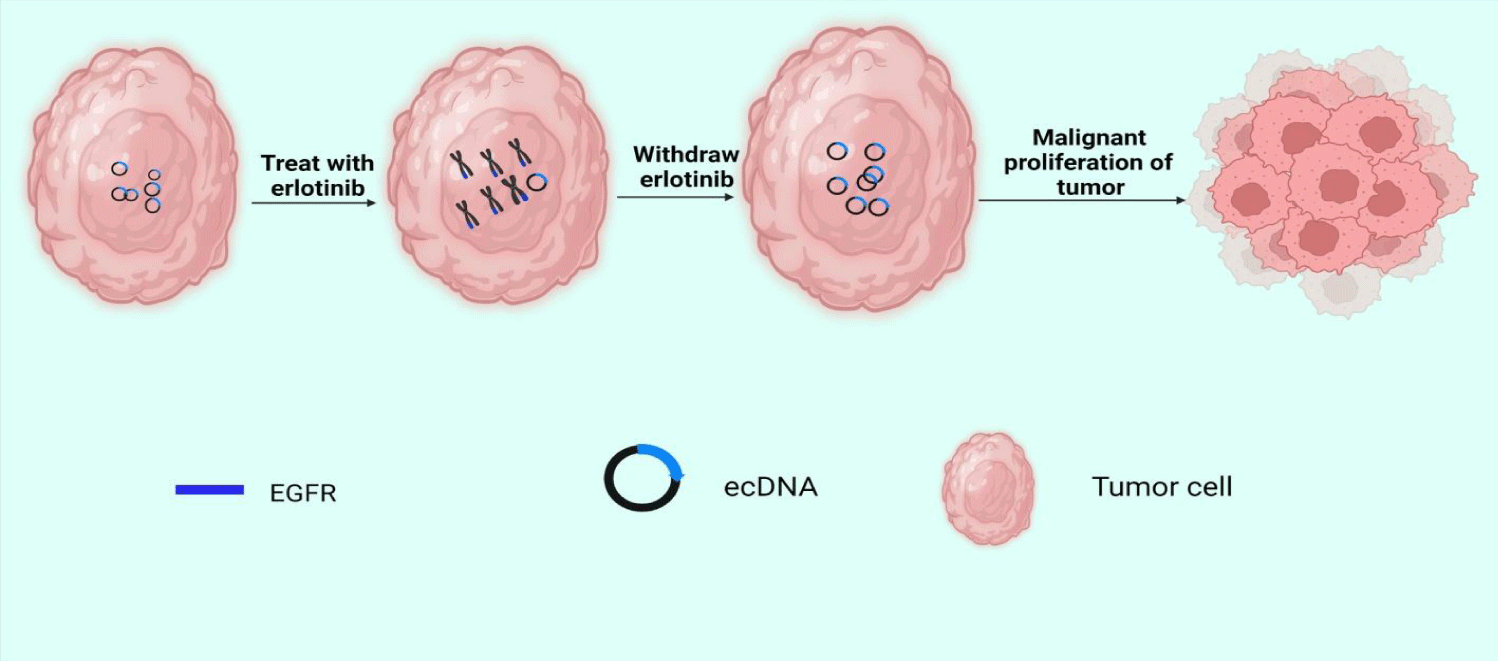
EcDNA and abnormal chromosomes are both carriers of oncogene proliferation in tumors, but the study of their relationship is still limited. Abnormal chromosomes are circular chromatids containing amplified oncogenes that provide selective growth advantages for tumor cells. Another study revealed amplicon's structure and expression pattern in acute myeloid leukemia and found the possibility of microsomes evolving towards circular chromosomes [60]. Scientist observed the same phenomenon in differentiated liposarcoma [61]. In addition, in some cases, clustered DMs can form micronucleus [27]. During the G1 phase of the mitotic interphase, DMs are located in the periphery of the nucleus. However, if cells are treated with low concentrations of DNA replication inhibitors, a subset of DMs aggregates to form micronucleus independent of chromosomes [62]. The formation of a micronucleus may reverse the malignant characteristics of the tumor to achieve the purpose of treatment. EcDNA can also be dynamically reintegrated into chromophores, leading to genome remodeling. In 2017, Turner, et al. [23] combined fluorescence in situ hybridization with second-generation sequencing to analyze the structure of the EGFRvIII amplicon. A key finding of their work was that after treatment with erlotinib, the ecDNA carrying EGFRvIII reintegrated into the chromosomes. When treatment with erlotinib is discontinued, ecDNA carrying EGFRvIII can be freed from the chromosome again (Figure 2). We think this amazing phenomenon may have something to do with tumor dormancy. Tumor dormancy refers to systemic or local tumor recurrence long after resection of the primary tumor. During this period, tumor growth is almost static, and patients live with tumors [63].
Research Methods for eccDNA
The research on eccDNA is still at the stage of identification and discovery. In order to identify and discover eccDNA, it is often necessary to evaluate the type and content of eccDNA in detected samples utilizing high-throughput sequencing. Since the content of eccDNA in cells and tissues is relatively low, the enrichment and purification of circular DNA should be carried out first before the study of eccDNA. At present, most methods for enrichment and purification of eccDNA rely on the special circular structure of eccDNA. In general, after the total DNA in cells or tissues to be detected is separated, the linearized DNA can be removed by a nuclease that can remove linear DNA without degrading circular DNA [16], and then the eccDNA can be amplified by rolling loop amplification [16,24]. After amplification, eccDNA can be quantified by double-terminal sequencing by analyzing the unique link sites of circular DNA (Circy-map) [64] or copy number variation (e.g., Amplicon Architect) [65]. Another method does not require rolling loop amplification to increase the copy number but uses Tn5 transposase to cut eccDNA [66]. This enzyme digestion method is easier to quantify than rolling loop amplification because it does not require amplification. The advantage of rolling loop amplification is that it is easy to fix, but the disadvantage is that the repeated amplification of DNA fragments will increase the signal-to-noise ratio [66].
Conclusion
Extragenomal circular DNA is a closed circular DNA molecule with single or double-stranded eukaryotes, which plays an important role in physiological and pathological processes. At present, researchers have reasonably speculated the formation process of eccDNA based on some phenomena and established relevant models. Further advances have been made in the function of eccDNA. Especially in 2019, Wu, et al. [3] and Morton, et al. [4] reported that ecDNA chromatin was more open than chromatids and carried functional enhancers, leading to the stronger transcriptional activity of oncogenes on ecDNA. These two discoveries have overturned the previous understanding that genes are located on chromosomes, pointing to a new direction and proposing a whole new idea for cancer treatment. However, there is still a lack of newer and more comprehensive methods to study the specific role of eccDNA in regulating physiological and pathological processes, and there are still many problems to be solved. For example, the specific molecular mechanism and post-processing of eccDNA synthesis and elimination have not been fully clarified. There is a limited amount of eccDNA in a cell, so what is the endpoint of replication for eccDNA? Can different types of eccDNA be converted to each other according to the needs of the cell? It is believed that eccDNA will be more and more widely used in the treatment of diseases and liquid biopsy in the future as these mysteries are revealed step by step.
Author Contribution
Safir Ullah Khan devised and designed the study. Munir Ullah khan performed surveys and analyzed data. The authors read and approved the manuscript.
Ethical Approval
This article does not contain any studies with human participants or animals performed by the author.
References
- Kujirai T, Kurumizaka H. Transcription through the nucleosome. Curr Opin Struct Biol. 2020 Apr;61:42-49. doi: 10.1016/j.sbi.2019.10.007. Epub 2019 Nov 29. PMID: 31790919.
- Hotta Y, Bassel A. Molecular size and circularity of DNA in cells of mammals and higher plants. Proc Natl Acad Sci U S A. 1965 Feb;53(2):356-62. doi: 10.1073/pnas.53.2.356. PMID: 14294069; PMCID: PMC219520.
- Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, Luebeck J, Rajkumar U, Diao Y, Li B, Zhang W, Jameson N, Corces MR, Granja JM, Chen X, Coruh C, Abnousi A, Houston J, Ye Z, Hu R, Yu M, Kim H, Law JA, Verhaak RGW, Hu M, Furnari FB, Chang HY, Ren B, Bafna V, Mischel PS. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature. 2019 Nov;575(7784):699-703. doi: 10.1038/s41586-019-1763-5. Epub 2019 Nov 20. PMID: 31748743; PMCID: PMC7094777.
- Morton AR, Dogan-Artun N, Faber ZJ, MacLeod G, Bartels CF, Piazza MS, Allan KC, Mack SC, Wang X, Gimple RC, Wu Q, Rubin BP, Shetty S, Angers S, Dirks PB, Sallari RC, Lupien M, Rich JN, Scacheri PC. Functional Enhancers Shape Extrachromosomal Oncogene Amplifications. Cell. 2019 Nov 27;179(6):1330-1341.e13. doi: 10.1016/j.cell.2019.10.039. Epub 2019 Nov 21. PMID: 31761532; PMCID: PMC7241652.
- Paulsen T, Kumar P, Koseoglu MM, Dutta A. Discoveries of Extrachromosomal Circles of DNA in Normal and Tumor Cells. Trends Genet. 2018 Apr;34(4):270-278. doi: 10.1016/j.tig.2017.12.010. Epub 2018 Jan 9. PMID: 29329720; PMCID: PMC5881399.
- Liao Z, Jiang W, Ye L, Li T, Yu X, Liu L. Classification of extrachromosomal circular DNA with a focus on the role of extrachromosomal DNA (ecDNA) in tumor heterogeneity and progression. Biochim Biophys Acta Rev Cancer. 2020 Aug;1874(1):188392. doi: 10.1016/j.bbcan.2020.188392. Epub 2020 Jul 28. PMID: 32735964.
- Cohen S, Houben A, Segal D. Extrachromosomal circular DNA derived from tandemly repeated genomic sequences in plants. Plant J. 2008 Mar;53(6):1027-34. doi: 10.1111/j.1365-313X.2007.03394.x. Epub 2007 Dec 15. PMID: 18088310.
- Ain Q, Schmeer C, Wengerodt D, Witte OW, Kretz A. Extrachromosomal Circular DNA: Current Knowledge and Implications for CNS Aging and Neurodegeneration. Int J Mol Sci. 2020 Apr 2;21(7):2477. doi: 10.3390/ijms21072477. PMID: 32252492; PMCID: PMC7177960.
- Dennin RH. Overlooked: Extrachromosomal DNA and Their Possible Impact on Whole Genome Sequencing. Malays J Med Sci. 2018 Mar;25(2):20-26. doi: 10.21315/mjms2018.25.2.3. Epub 2018 Apr 27. PMID: 30918452; PMCID: PMC6422590.
- Verhaak RGW, Bafna V, Mischel PS. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat Rev Cancer. 2019 May;19(5):283-288. doi: 10.1038/s41568-019-0128-6. PMID: 30872802; PMCID: PMC7168519.
- Yan Y, Guo G, Huang J. Current understanding of ex-trachromosomal circular DNA in cancer pathogenesis and thera-peutic resistance. J Hematol Oncol. 2020;13(1):124. doi: 10.1186/s13045-020-00960-9.
- Khan SU, Khan MU. The mechanism of mammalian mitochondrial quality control system. Journal of Chemistry and Nutritional Biochemistry. 2021;59-69. doi: 10.48185/jcnb.v2i2.387.
- Storlazzi CT, Fioretos T, Surace C. MYC-containing double minutes in hematologic malignancies: evidence in favor of the episome model and exclusion of MYC as the target gene. Hum Mol Genet. 2006;15(6):933-42. doi: 10.1093/hmg/ddl010.
- Schmidt H, Taubert H, Lange H, Kriese K, Schmitt WD, Hoffmann S, Bartel F, Hauptmann S. Small polydispersed circular DNA contains strains of mobile genetic elements and occurs more frequently in permanent cell lines of malignant tumors than in normal lymphocytes. Oncol Rep. 2009 Aug;22(2):393-400. PMID: 19578782.
- Dennin RH, Wo JE. DNA sequences homologous to hepatitis C virus (HCV) in the extrachromosomal circular DNA in peripheral blood mononuclear cells of HCV-negative subjects. J Zhejiang Univ Sci B. 2019 Aug.;20(8):637-646. doi: 10.1631/jzus.B1800453. PMID: 31273961; PMCID: PMC6656560.
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Quail MA, Burton J, Swerdlow H, Carter NP, Morsberger LA, Iacobuzio-Donahue C, Follows GA, Green AR, Flanagan AM, Stratton MR, Futreal PA, Campbell PJ. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011 Jan 7;144(1):27-40. doi: 10.1016/j.cell.2010.11.055. PMID: 21215367; PMCID: PMC3065307.
- Kumar P, Dillon LW, Shibata Y, Jazaeri AA, Jones DR, Dutta A. Normal and Cancerous Tissues Release Extrachromosomal Circular DNA (eccDNA) into the Circulation. Mol Cancer Res. 2017 Sep;15(9):1197-1205. doi: 10.1158/1541-7786.MCR-17-0095. Epub 2017 May 26. PMID: 28550083; PMCID: PMC5581709.
- Wang T, Zhang H, Zhou Y, Shi J. Extrachromosomal circular DNA: a new potential role in cancer progression. J Transl Med. 2021 Jun 10;19(1):257. doi: 10.1186/s12967-021-02927-x. PMID: 34112178; PMCID: PMC8194206.
- Cox D, Yuncken C, Spriggs AI. Minute chromatin bodies in malignant tumours of childhood. Lancet. 1965 Jul 10;1(7402):55-8. doi: 10.1016/s0140-6736(65)90131-5. PMID: 14304929.
- Dahm R. Discovering DNA: Friedrich Miescher and the early years of nucleic acid research. Hum Genet. 2008 Jan;122(6):565-81. doi: 10.1007/s00439-007-0433-0. Epub 2007 Sep 28. PMID: 17901982.
- Khan S, Khan M. Molecular developments in cell models of fatty liver disease. DYSONA-Life Science. 2022;1:16-29. doi: 10.30493/DLS.2022.325915.
- Shibata Y, Kumar P, Layer R, Willcox S, Gagan JR, Griffith JD, Dutta A. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012 Apr 6;336(6077):82-6. doi: 10.1126/science.1213307. Epub 2012 Mar 8. Erratum in: Science. 2012 Jun 22;336(6088):1506. PMID: 22403181; PMCID: PMC3703515.
- Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, Li B, Arden K, Ren B, Nathanson DA, Kornblum HI, Taylor MD, Kaushal S, Cavenee WK, Wechsler-Reya R, Furnari FB, Vandenberg SR, Rao PN, Wahl GM, Bafna V, Mischel PS. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017 Mar 2;543(7643):122-125. doi: 10.1038/nature21356. Epub 2017 Feb 8. PMID: 28178237; PMCID: PMC5334176.
- Moller HD, Mohiyuddin M, Prada-Luengo I, Sailani MR, Halling JF, Plomgaard P, Maretty L, Hansen AJ, Snyder MP, Pilegaard H, Lam HYK, Regenberg B. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat Commun. 2018 Mar 14;9(1):1069. doi: 10.1038/s41467-018-03369-8. PMID: 29540679; PMCID: PMC5852086.
- Paulsen T, Shibata Y, Kumar P, Dillon L, Dutta A. Small extrachromosomal circular DNAs, microDNA, produce short regulatory RNAs that suppress gene expression independent of canonical promoters. Nucleic Acids Res. 2019 May 21;47(9):4586-4596. doi: 10.1093/nar/gkz155. PMID: 30828735; PMCID: PMC6511871.
- Bailey C, Shoura MJ, Mischel PS, Swanton C. Extrachromosomal DNA-relieving heredity constraints, accelerating tumour evolution. Ann Oncol. 2020 Jul;31(7):884-893. doi: 10.1016/j.annonc.2020.03.303. Epub 2020 Apr 7. PMID: 32275948.
- Ruiz JC, Choi KH, von Hoff DD, Roninson IB, Wahl GM. Autonomously replicating episomes contain mdr1 genes in a multidrug-resistant human cell line. Mol Cell Biol. 1989 Jan;9(1):109-15. doi: 10.1128/mcb.9.1.109-115.1989. PMID: 2648129; PMCID: PMC362151.
- Khan SU. Therapeutic application of genetically engineered ribosome-inactivating toxin proteins for cancer. J Biomed Res Environ Sci. 2021;2(12):1216-1228. doi: 10.37871/jbres1375.
- Dillon LW, Kumar P, Shibata Y, Wang YH, Willcox S, Griffith JD, Pommier Y, Takeda S, Dutta A. Production of Extrachromosomal MicroDNAs Is Linked to Mismatch Repair Pathways and Transcriptional Activity. Cell Rep. 2015 Jun 23;11(11):1749-59. doi: 10.1016/j.celrep.2015.05.020. Epub 2015 Jun 4. PMID: 26051933; PMCID: PMC4481157.
- Chiu RWK, Dutta A, Henssen AG, Dennis LYM, Mischel P, Regenberg B. What is extra-chromosomal circular DNA and what does it do. Clin Chem. 2020;66(6):754-9. doi: 10.1093/clinchem/hvaa096.
- Gu X, Yu J, Chai P, Ge S, Fan X. Novel insights into extrachromosomal DNA: redefining the onco-drivers of tumor progression. J Exp Clin Cancer Res. 2020 Oct 12;39(1):215. doi: 10.1186/s13046-020-01726-4. PMID: 33046109; PMCID: PMC7552444.
- Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005 Apr;15(2):188-96. doi: 10.1016/j.sbi.2005.03.006. PMID: 15837178.
- Alisch RS, Barwick BG, Chopra P, Myrick LK, Satten GA, Conneely KN, Warren ST. Age-associated DNA methylation in pediatric populations. Genome Res. 2012 Apr;22(4):623-32. doi: 10.1101/gr.125187.111. Epub 2012 Feb 1. PMID: 22300631; PMCID: PMC3317145.
- Neurohr GE, Terry RL, Sandikci A, Zou K, Li H, Amon A. Deregulation of the G1/S-phase transition is the proximal cause of mortality in old yeast mother cells. Genes Dev. 2018 Aug 1;32(15-16):1075-1084. doi: 10.1101/gad.312140.118. Epub 2018 Jul 24. PMID: 30042134; PMCID: PMC6075151.
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997 Dec 26;91(7):1033-42. doi: 10.1016/s0092-8674(00)80493-6. PMID: 9428525.
- Eickbush TH, Eickbush DG. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007 Feb;175(2):477-85. doi: 10.1534/genetics.107.071399. PMID: 17322354; PMCID: PMC1800602.
- Horigome C, Kobayashi T. Rejuvenation of ribosomal RNA gene repeats at the nuclear pore. Curr Genet. 2020 Feb;66(1):7-13. doi: 10.1007/s00294-019-01024-3. Epub 2019 Aug 7. PMID: 31392389.
- Ganley AR, Ide S, Saka K, Kobayashi T. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol Cell. 2009 Sep 11;35(5):683-93. doi: 10.1016/j.molcel.2009.07.012. PMID: 19748361.
- Khan SU, Khan MU. Review on gene regulation: DNA-protein and protein-protein interactions and their regulatory elements. Journal of Chemistry and Nutritional Biochemistry. 2021;2(2):35-45. doi: 10.48185/jcnb.v2i2.378.
- Mansisidor A, Molinar T Jr, Srivastava P, Dartis DD, Pino Delgado A, Blitzblau HG, Klein H, Hochwagen A. Genomic Copy-Number Loss Is Rescued by Self-Limiting Production of DNA Circles. Mol Cell. 2018 Nov 1;72(3):583-593.e4. doi: 10.1016/j.molcel.2018.08.036. Epub 2018 Oct 4. PMID: 30293780; PMCID: PMC6214758.
- Burkhalter MD, Sogo JM. rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol Cell. 2004 Aug 13;15(3):409-21. doi: 10.1016/j.molcel.2004.06.024. PMID: 15304221.
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999 Apr;3(4):447-55. doi: 10.1016/s1097-2765(00)80472-4. PMID: 10230397.
- Morlot S, Song J, Léger-Silvestre I, Matifas A, Gadal O, Charvin G. Excessive rDNA Transcription Drives the Disruption in Nuclear Homeostasis during Entry into Senescence in Budding Yeast. Cell Rep. 2019 Jul 9;28(2):408-422.e4. doi: 10.1016/j.celrep.2019.06.032. PMID: 31291577.
- Del Zotto G, Principi E, Antonini F, Baratto S, Panicucci C, Bruno C, Raffaghello L. Comprehensive Phenotyping of Peripheral Blood T Lymphocytes in Healthy Mice. Cytometry A. 2021 Mar;99(3):243-250. doi: 10.1002/cyto.a.24246. Epub 2020 Nov 11. PMID: 33098601.
- Jones JM, Gellert M. The taming of a transposon: V(D)J recombination and the immune system. Immunol Rev. 2004 Aug;200:233-48. doi: 10.1111/j.0105-2896.2004.00168.x. PMID: 15242409.
- Hazenberg MD, Verschuren MC, Hamann D, Miedema F, van Dongen JJ. T cell receptor excision circles as markers for recent thymic emigrants: basic aspects, technical approach, and guidelines for interpretation. J Mol Med (Berl). 2001 Nov;79(11):631-40. doi: 10.1007/s001090100271. PMID: 11715066.
- Jung D, Alt FW. Unraveling V(D)J recombination; insights into gene regulation. Cell. 2004 Jan 23;116(2):299-311. doi: 10.1016/s0092-8674(04)00039-x. PMID: 14744439.
- Sun W, Quan C, Huang Y, Ji W, Yu L, Li X, Zhang Y, Zheng Z, Zou H, Li Q, Xu P, Feng Y, Li L, Zhang Y, Cui Y, Jia X, Meng X, Zhang C, Jin Y, Bai J, Yu J, Yu Y, Yang J, Fu S. Constitutive ERK1/2 activation contributes to production of double minute chromosomes in tumour cells. J Pathol. 2015 Jan;235(1):14-24. doi: 10.1002/path.4439. Epub 2014 Nov 6. PMID: 25214430; PMCID: PMC4280677.
- Kuttler F, Mai S. Formation of non-random extrachromosomal elements during development, differentiation and oncogenesis. Semin Cancer Biol. 2007 Feb;17(1):56-64. doi: 10.1016/j.semcancer.2006.10.007. Epub 2006 Oct 26. PMID: 17116402.
- Cai M, Zhang H, Hou L, Gao W, Song Y, Cui X, Li C, Guan R, Ma J, Wang X, Han Y, Lv Y, Chen F, Wang P, Meng X, Fu S. Inhibiting homologous recombination decreases extrachromosomal amplification but has no effect on intrachromosomal amplification in methotrexate-resistant colon cancer cells. Int J Cancer. 2019 Mar 1;144(5):1037-1048. doi: 10.1002/ijc.31781. Epub 2018 Sep 29. PMID: 30070702; PMCID: PMC6586039.
- Serana F, Chiarini M, Zanotti C, Sottini A, Bertoli D, Bosio A, Caimi L, Imberti L. Use of V(D)J recombination excision circles to identify T- and B-cell defects and to monitor the treatment in primary and acquired immunodeficiencies. J Transl Med. 2013 May 9;11:119. doi: 10.1186/1479-5876-11-119. PMID: 23656963; PMCID: PMC3666889.
- Gaballa A, Clave E, Uhlin M, Toubert A, Arruda LCM. Evaluating Thymic Function After Human Hematopoietic Stem Cell Transplantation in the Personalized Medicine Era. Front Immunol. 2020 Jul 31;11:1341. doi: 10.3389/fimmu.2020.01341. PMID: 32849495; PMCID: PMC7412601.
- Khan SU, Khan MU. Recent Developments and Applications of Single-Cell RNA Sequencing Technology in Cell Classification. J Biomed Res Environ Sci. 2021 Dec 29;2(12):1283-1290. doi: 10.37871/jbres1383.
- Cohen S, Yacobi K, Segal D. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 2003 Jun;13(6A):1133-45. doi: 10.1101/gr.907603. PMID: 12799349; PMCID: PMC403641.
- Cohen S, Mechali M. A novel cell-free system reveals a mechanism of circular DNA formation from tandem repeats. Nucleic Acids Res. 2001 Jun 15;29(12):2542-8. doi: 10.1093/nar/29.12.2542. PMID: 11410662; PMCID: PMC55730.
- Tanaka H, Watanabe T. Mechanisms Underlying Recurrent Genomic Amplification in Human Cancers. Trends Cancer. 2020 Jun;6(6):462-477. doi: 10.1016/j.trecan.2020.02.019. Epub 2020 Mar 24. PMID: 32383436; PMCID: PMC7285850.
- Shimizu N, Shingaki K, Kaneko-Sasaguri Y, Hashizume T, Kanda T. When, where and how the bridge breaks: anaphase bridge breakage plays a crucial role in gene amplification and HSR generation. Exp Cell Res. 2005 Jan 15;302(2):233-43. doi: 10.1016/j.yexcr.2004.09.001. PMID: 15561104.
- Khan SU, Khan MU. The role of amino acid metabolic reprogramming in tumor development and immunotherapy. Biochemistry and Molecular Biology. 2022;7(1):6-12. doi: 10.11648/j.bmb.20220701.12.
- Brewer BJ, Payen C, Raghuraman MK, Dunham MJ. Origin-dependent inverted-repeat amplification: a replication-based model for generating palindromic amplicons. PLoS Genet. 2011 Mar;7(3):e1002016. doi: 10.1371/journal.pgen.1002016. Epub 2011 Mar 17. PMID: 21437266; PMCID: PMC3060070.
- L Abbate A, Tolomeo D, Cifola I, Severgnini M, Turchiano A, Augello B, Squeo G, D Addabbo P, Traversa D, Daniele G, Lonoce A, Pafundi M, Carella M, Palumbo O, Dolnik A, Muehlematter D, Schoumans J, Van Roy N, De Bellis G, Martinelli G, Merla G, Bullinger L, Haferlach C, Storlazzi CT. MYC-containing amplicons in acute myeloid leukemia: genomic structures, evolution, and transcriptional consequences. Leukemia. 2018 Oct;32(10):2152-2166. doi: 10.1038/s41375-018-0033-0. Epub 2018 Feb 22. Erratum in: Leukemia. 2018 Jul 9;: PMID: 29467491; PMCID: PMC6170393.
- Venkatesan S, Swanton C. Tumor Evolutionary Principles: How Intratumor Heterogeneity Influences Cancer Treatment and Outcome. Am Soc Clin Oncol Educ Book. 2016;35:e141-9. doi: 10.1200/EDBK_158930. PMID: 27249716.
- McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell. 2017 Feb 9;168(4):613-628. doi: 10.1016/j.cell.2017.01.018. PMID: 28187284.
- Sacristan C, Ahmad MUD, Keller J, Fermie J, Groenewold V, Tromer E, Fish A, Melero R, Carazo JM, Klumperman J, Musacchio A, Perrakis A, Kops GJ. Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis. Nat Cell Biol. 2018 Jul;20(7):800-810. doi: 10.1038/s41556-018-0130-3. Epub 2018 Jun 18. PMID: 29915359; PMCID: PMC6485389.
- Amirouchene-Angelozzi N, Swanton C, Bardelli A. Tumor Evolution as a Therapeutic Target. Cancer Discov. 2017 Jul 20. doi: 10.1158/2159-8290.CD-17-0343. Epub ahead of print. PMID: 28729406.
- Kim H, Nguyen NP, Turner K, Wu S, Gujar AD, Luebeck J, Liu J, Deshpande V, Rajkumar U, Namburi S, Amin SB, Yi E, Menghi F, Schulte JH, Henssen AG, Chang HY, Beck CR, Mischel PS, Bafna V, Verhaak RGW. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet. 2020 Sep;52(9):891-897. doi: 10.1038/s41588-020-0678-2. Epub 2020 Aug 17. PMID: 32807987; PMCID: PMC7484012.
- Mitsuda SH, Shimizu N. Epigenetic Repeat-Induced Gene Silencing in the Chromosomal and Extrachromosomal Contexts in Human Cells. PLoS One. 2016 Aug 15;11(8):e0161288. doi: 10.1371/journal.pone.0161288. PMID: 27525955; PMCID: PMC4985131.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.