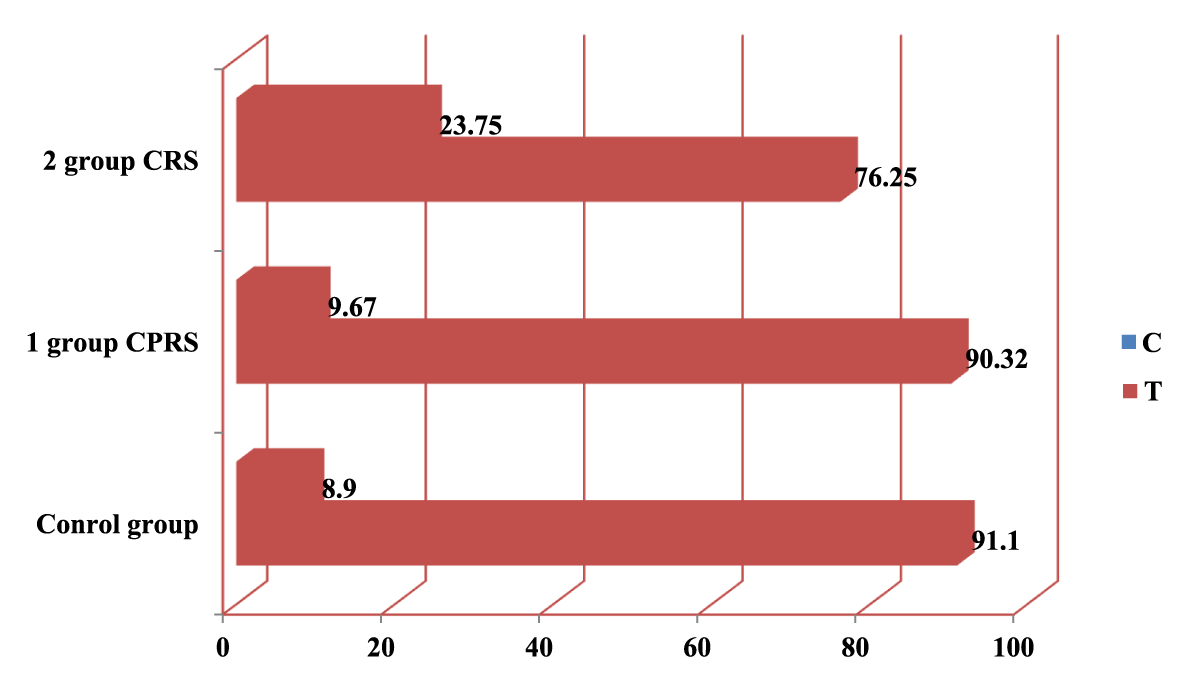> Medicine Group. 2021 June 14;2(6):486-492. doi: 10.37871/jbres1264.
Frequency Analysis Results Distribution of C589t Rs2243250 Polymorphism in Il4 Gene Among Patients with Chronic Rhinosinusitis
Djuraev JA1*, Khasanov US2, Vokhidov UN3 and Botirov AJ4
2Doctor of Medical Sciences, Professor, Head of the Department of Otolaryngology and Dentistry, Tashkent Medical Academy, Republic of Uzbekistan
3Doctor of Medical Sciences, Associate Professor of the Department of Otorhinolaryngology, Tashkent State Dental Institute, Republic of Uzbekistan
4Department of Otolaryngology and Dentistry, Tashkent Medical Academy, Republic of Uzbekistan
- Polyp
- Paranasal sinuses
- Interleukin
- Cytokine
- Inflammatory process
Abstract
The fairly widespread prevalence of CRSwNP along with the lack of remedies for curing the disease, a variety of hypotheses of etiology dictate the need for further study of all links in the pathogenesis and clinical features of the course of the disease. In the coming century of “biological medicine”, the availability of high technologies of medical genetics makes it possible to reveal the individual characteristics of the most important regulatory systems of the body, which opens up new prospects for studying the etiology and pathogenesis of CRSwNP. In the tissues of polyps and intranasal secretions, an increase in the concentration of various inflammatory mediators, in particular interleukins, is observed due to an increase in their de novo synthesis by effector cells. Particular importance is attached to an increase in the concentration of cytokines involved in the development, recruitment and activation of eosinophils (IL-4, IL-12, IL-13, GM-CSF), the main pro-inflammatory (IL-1, IL-2, TNF-a, IL- 10), regulatory cytokines (IL-10, TLR2B), contributing to the chronicity of the inflammatory process in the nasal cavity.
Introduction
Diseases of the paranasal sinuses are among the most common pathologies in otorhinolaryngology, which is facilitated by the modern environmental situation, the widespread prevalence of allergic and viral respiratory diseases, and a decrease in local and general immunity. All researchers agree that in recent years in the world there has been a tendency towards an increase in the incidence of chronic sinusitis, including Chronic Polypous Rhinosinusitis (CRSwNP) [1].
Epidemiological studies of CRSwNP in Europe, which were carried out with an interval of 8 years, indicate that at selected time intervals in each specific region, the prevalence of the disease does not change significantly [2]. For a number of reasons (environmental conditions, social and drug load, changes in the functional indicators of the most important homeostatic systems of the human body, etc.), it is not necessary to expect a decrease in the incidence of CRSwNP [3]. The stability of CRSwNP incidence rates, regardless of regional characteristics or other external factors, is considered by leading otolaryngologists to be the basis for a more detailed study of the causes of this nosology [4], first of all - genetic predisposition to CRSwNP development. Many facts speak in favor of the genetic hypothesis of CRSwNP development. It has been proven that the risk of CRSwNP in the presence of polyposis heredity is 25 times higher, with a heterozygous carriage of the MZ phenotype (deficiency of alpha-1 antitrypsinase) - 4 times, with a dry type of earwax - 3 times [5,6]; revealed changes in the karyotypes of peripheral blood cells in patients with CRSwNP [7]. Since chromosomal polymorphism can determine individual sensitivity to the occurrence of any disease, i.e. individual response of the organism to a damaging factor, persons with karyotype variants that differ from the norm are at risk of developing certain diseases that depend on hypo-, hyper, or norm-sensitivity of the hereditary apparatus [8].
Numerous studies of the last decade have demonstrated the dependence of the immune response on allelic polymorphism of cytokine genes. The result of such works in vitro is the identification of individual alleles of genes associated with increased or decreased production of the corresponding cytokine [9]. The data obtained to date suggest that polymorphic cytokine genes are able to take an active part in the formation of a specific immune response to pathological conditions in humans [10]. Individual allelic variants can be associated with the level of production of the corresponding protein, which also affects the course of the disease and the development of a number of complications. However, it remains unclear which mutations and which cytokines are of decisive importance in the development of individual diseases. Therefore, a promising area of molecular genetic research is the study of the contribution of specific alleles to the propensity to infection in the development of pathology [11,12].
The modern stage in the development of cytology, histology and clinical anatomy, as well as progress in diagnostic technologies have led to the concept of the nasal cavity as a complex morphofunctional system [13]. The modern knowledge obtained in the process of scientific research about anatomy, histology and physiology, as well as the morphogenesis of various pathological processes in the nasal cavity and paranasal sinuses, significantly expanded the idea of the functional significance of these structures in the adaptive capabilities of the nasal cavity to breathing conditions, their role in the respiratory system as a whole [14].
Modern histological and clinical-functional studies made it possible to state the growth of chronic diseases of the mucous membrane of the nasal cavity and paranasal sinuses, the formation of various endonasal formations [15]. This is due to the deterioration of the ecological and social situation, an increase in the virulence of the microbial flora, a change in its composition and resistance to antibacterial drugs. In the pathogenesis of diseases of the ENT organs, in addition to the infectious agent, the leading role belongs to the immune system of the mucous membranes of the nose and pharynx, as well as the general reactions of humoral and cellular immunity [16].
The body’s resistance to exo- and endogenous pathological factors is largely associated with the ability to quickly adapt to changing environmental conditions. The mucous membrane of the nasal cavity serves as the first protective barrier where local immunity reactions take place.
It is known that in many organs and tissues, including the mucous membrane of the nasal cavity of the human body, there are neuroendocrine cells related to the link of autonomous regulation of organs. Moreover, the structural and functional features of neuroendocrine cells and their bioamine profile in patients with signs of polyposis rhinosinusitis have not been practically studied. Therefore, the study of the morphophysiological organization of the mucous membrane of the nasal cavity in humans is an urgent problem of modern cytology, histology and cell biology [17].
Considering the above, we conducted a study of the genetic polymorphism of cytokine genes in patients with CRSwNP, the results of which demonstrate genetically determined features of the immune response that contribute to the development of CRSwNP, as well as determining some of the clinical features of the disease.
The aim of this work was to study the influence of some genetic factors on the development and course of chronic polypous rhinosinusitis.
Material and Methods
Molecular genetic studies were carried out in the Department of Molecular Medicine and Cell Technologies.
This part of the work consisted of several stages:
• Blood sampling
• Isolation of DNA from peripheral blood lymphocytes
• Carrying out PCR
In the course of the work, 4 polymorphic variants of genes were investigated (Table 1).
| Table 1: List of studied gene polymorphisms. | |||
| Gene (abbreviation) | Polymorphism | International code | A source |
| Interleukin (IL)-4 | C589T | rs 2243250 | Morrison N.A |
Note:
• Vickers MA, Green FR, Terry C, Mayosi BM, Julier C, Lathrop M, Ratcliffe PJ, Watkins HC, Keavney B. Genotype at a promoter polymorphism of the interleukin-6 gene is associated with baseline levels of plasma C-reactive protein. Cardiovasc Res. 2002 Mar;53(4):1029-34. doi: 10.1016/s0008-6363(01)00534-x. PMID: 11922913.
• Braun N, Michel U, Ernst BP, Metzner R, Bitsch A, Weber F, Rieckmann P. Gene polymorphism at position -308 of the tumor-necrosis-factor-alpha (TNF-alpha) in multiple sclerosis and it’s influence on the regulation of TNF-alpha production. Neurosci Lett. 1996 Sep 6;215(2):75-8. PMID: 8887999.
• Vincenti V, Cassano C, Rocchi M, Persico G. Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3. Circulation. 1996 Apr 15;93(8):1493-5. doi: 10.1161/01.cir.93.8.1493. PMID: 8608615.
The analysis of the associations of the IL4 - C589T gene polymorphism was carried out using a case-control model (case-control, comparison of two samples). The sample “case” was formed from 71 patients with CRSwNP and CRS. The average age of the patients was 41.2. All examined patients were divided into 3 groups:
1. Patients with CRS (group I; n = 71);
2. HPRS
3. Other forms of CRS; control group (group II; n = 50).
Genomic DNA preparations, both isolated independently and stored in the DNA bank of the RSNPMC Hematology of the Ministry of Health of the Republic of Uzbekistan, were used as material for the control sample. The control group consisted of 50 healthy unrelated donors (Uzbek nationality), matched by sex and age to the examined group of patients (p > 0.05), and had no history of CRS pathology.
Genetic research and analysis of the data obtained were carried out according to the principles of GRIPS in order to increase transparency and the quality of risk prediction [10].
The AmpliPrime RIBO-prep reagent kit (AmpliSens, Russia) was used to isolate DNA from peripheral blood. The isolated DNA concentrations were measured on a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, United States) at a wavelength of A260/280 nm. The purity of all samples of the isolated DNA preparation, determined by the ratio A260/280, was 1.7/1.8.
The search for gene sequences for the selection of oligoprimers was carried out in the GenBank NCBI (http://www.ncbi.nlm.nih.gov/GenBank). Nucleotide sequences and evaluations of oligoprimer characteristics were performed using the Oligo v.6.31 software (Molecular Biology Insights Inc., USA). To carry out standard PCR, we used a Gene Pak® PCR MasterMix Core DNA amplification kit (IsoGene Lab. Ltd., Russia).
The following reagents and enzymes were used in the work: acrylamide, bis-acrylamide, EDTA, 5% glycerol solution, proteinase K (Sigma, USA), TEMED, sodium dodecyl sulfate, Tris-HCL (Serva, Germany), 2 -mercaptoethanol (“Ferax”, Germany), deoxynucleotide triphosphates, dideoxynucleotide triphosphates, Triton X100, magnesium chloride, sodium chloride, ammonium sulfate, ammonium persulfate, thermostable DNA polymerase Thermus aquaticus (NPO “Biomereotide” , Novosibirsk), test systems by OOO NPF Litekh (Moscow) and OOO InterLabService (Moscow).
Commercial standard kits
Kit for DNA / RNA isolation “Ribo-prep” LLC InterLabService (Moscow), a kit for testing the polymorphism of the rs2243250 gene manufactured by OOO NPF Litekh (Moscow).
Laboratory equipment
For molecular genetic studies, the following equipment was used: Applied Biosystems 2720 (USA) and СG1-96 thermal cyclers (Corbett Research, QUAGEN Germany), and RotorGeneQ (QUAGEN Germany), laminar box (Germany), centrifuges (Eppendorf, Hittich, Germany) , vortex (Eppendorf, Germany), thermostats, spectrophotometer NanoDrop 2000 “Thermo Scientific” (USA), power supply (DNA-Technology, Russia), UV transilluminator with built-in digital camera, automatic pipettes (Sartorius, Finland) and etc.
Extraction of genomic DNA from peripheral blood lymphocytes
To isolate DNA from peripheral blood lymphocytes, we used a modified phenol-chloroform extraction method and an RNA/DNA-sorb kit from InterLabService LLC (Russia). When sampling the biomaterial, standard vacuum tubes Vacutainer Becton Dickinson International (USA) with EDTA were used.
Isolation of lymphocyte nuclei and subsequent DNA was carried out in accordance with the method proposed by Sambrook J, with some modifications. Citrated blood was mixed with an equal volume of buffer (4°C) containing: 0.32 M sucrose; 5 mM MgCl2; 1% Triton X-100; 0.01M Tris-HCl pH 7.5. This mixture was centrifuged at 3000 rpm at 4°C. The nuclear pellet was resuspended in 400 µl of proteinase K buffer, composition: 10 mM Tris-HCl, pH 10.5; 0.5M NaCl; 1mM EDTA. SDS (Serva, Germany) was added to a final concentration of 0.5% and incubated in the presence of proteinase K (Serva, Germany or Sigma, USA), at a concentration of 250 μg/ml for 16 hours at 37°C. 400 μL of buffered phenol was added, gently mixed for 10 min, and centrifuged for 5 min at 5000 rpm. Then, the upper phase was transferred to another test tube, and 400 μL of a phenol: chloroform mixture (1:1) was added. Stirred for 5 min, centrifuged. Phenol was extracted from the upper aqueous phase with an equal volume of chloroform. To the DNA solution were added sequentially 40 μl of 3M sodium acetate and 800 μl of chilled 96% ethanol. The mixture was stirred and centrifuged for 15 min at 14000 rpm, the precipitate was washed with 1 ml of 70% ethanol. It was centrifuged again, the precipitate was dried and DNA was dissolved in TE buffer (10 mM Tris-HCl pH 7.4; 1 mM EDTA, pH 8.0) for 12 hours at room temperature. The concentration and purity of the isolated DNA were measured on a NanoDrop 2000 spectrophotometer (USA) at a wavelength of A260/280 nm. The purity of the isolated DNA samples, determined by the ratio A260/280, was 1.7/1.8. This indicates a very low content of contaminating proteins or other macromolecules in solutions of isolated DNA, and these samples can be used in PCR without additional purification. Genomic DNA solution 1 mg/ml, equivalent to 20 p.u. DNA was stored in TE at -20°C.
Mathematical methods of analysis
Evaluation of the deviation of the distributions of genotypes of the studied DNA polymorphisms from the canonical Hardy-Weinberg distribution was carried out using the computer program for the analysis of genetic data “GenePop” (“Genetics of Population”), available on the Internet (http://wbiomed.curtin.edu.au/genepop).
The allele frequencies of the studied genes were calculated using the following formula:
p = (2np + npq)/2N, where N is the sample size; np is the number of homozygotes for allele p; npq is the number of heterozygotes.
Actual (observed) heterozygosity:
Нobs = No/N, where No is the number of heterozygotes.
Theoretical (expected) heterozygosity:
Hexp = 1 - Σpi2, where pi is the frequency of the i allele.
i = 1
The coefficient of deviation of the actual heterozygosity from the theoretical was calculated using the following formula:
F = (Hexp –Hobs)/Hexp.
In order to determine the effectiveness of each genetic marker, the Sensitivity (SE), Specificity (SP) and AUC (area under curve) were calculated.
The predictive value was determined as follows: if the AUC indicator is <0.5, then the marker is random; 0.5
When comparing the frequencies of alleles and genotypes in the groups of patients and controls, the χ2 criterion was used. For contingency tables 2 × 2, the χ2 test was used with Yates’ correction for continuity if the frequency in at least one cell of the table was less than or equal to 5. Differences were considered statistically significant at p < 0.05.
The degree of associations was assessed in terms of the Odds Ratio (OR) and its 95% confidence interval (95% CI), using the formula:
OR = (a x d)/(b x c), where a is the frequency of the allele (genotype) in the sample of patients, b is the frequency of the allele (genotype) in the control sample, c is the sum of the frequencies of the remaining alleles (genotypes) in the sample of patients, d is the sum of the frequencies of the remaining alleles (genotypes) in the control sample. OR = 1 indicated no association. OR>1 was considered as a positive association of the disease with a trait (increased risk factor), OR<1 - as a negative association (reduced risk factor).
The software package “OpenEpi 2009, Version 2.3” was used as a calculation tool.
Results and Discussion
The study of the frequencies of detection of alleles and genotypes of the C589T rs2243250 polymorphism in the IL4 gene showed the presence of differences in their distribution between 1-2 and control groups (Table 2).
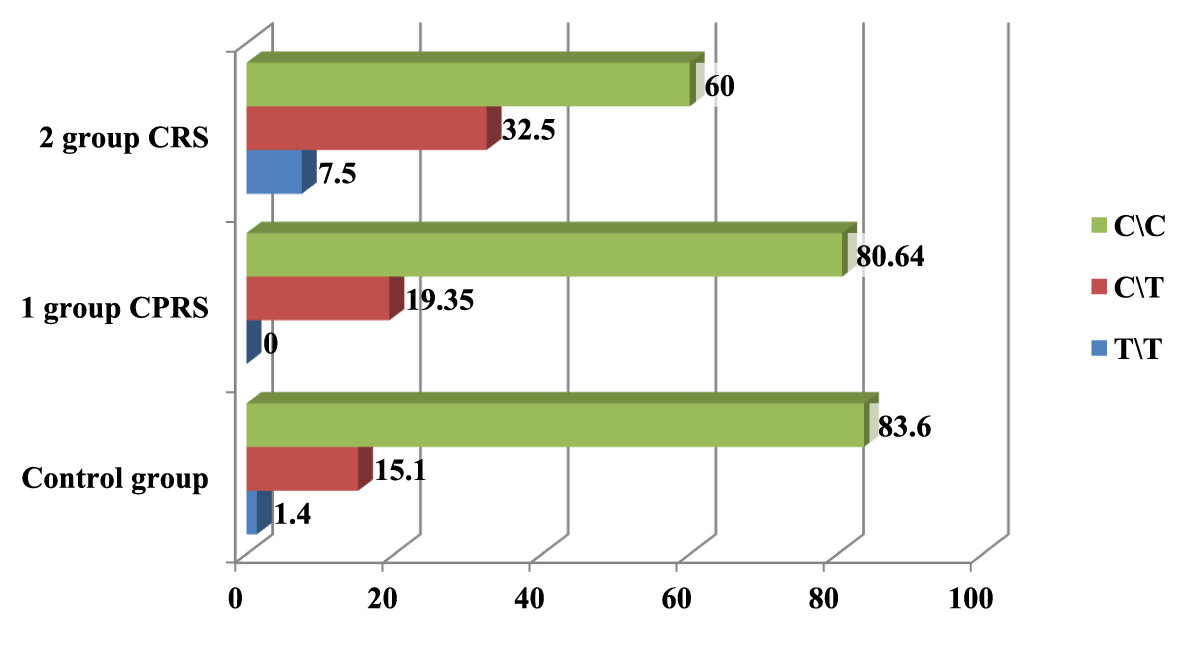
In the course of the study, it was possible to establish the frequency of detection of the C allele, which was 9.3 times higher than the frequency of detection of the T allele in group 1, 3.2 in group 2, and 10.1 times in the control group. The C/C genotype in the 1st group, in comparison with the C/T and T/T genotypes, was detected more often 4.16 times, respectively, in the 2nd group, 1.84 times, and in the population sample, 5.53 times, respectively (Table 2, Figures 1,2).
| Table 2: Frequency of distribution of alleles and genotypes of C589T rs2243250 polymorphism in the IL4 gene in groups of patients with CRSWNP and CRS. | |||||||||||
| № | Group | Allele frequency | Genotype distribution frequency | ||||||||
| C | T | C\C | C\T | T\T | |||||||
| n | % | n | % | n | % | n | % | n | % | ||
| 1 | CRSWNP n = 31 | 56 | 90.32 | 6 | 9.67 | 25 | 80.64 | 6 | 19.35 | 0 | 0 |
| 2 | CRS n = 40 | 61 | 76.25 | 19 | 23.75 | 24 | 60 | 13 | 32.5 | 3 | 7.5 |
| 3 | Control group n = 73 | 133 | 91.1 | 13 | 8.9 | 61 | 83.6 | 11 | 15.1 | 1 | 1.4 |
The results of a comparative analysis of the frequencies of detection of alleles and genotypes of the C589T rs2243250 polymorphism in the IL4 gene of group 1 and in the population sample are presented in table 3.
| Table 3: Differences in the frequency of allelic and genotypic variants polymorphism C589T rs2243250 in the IL4 gene in patients with CRSWNP and among conventionally healthy individuals. | ||||||||||
| Alleles and genotypes | Number of examined alleles and genotypes | Xi2 | p | RR | + 95%CI | OR | +95%CI | |||
| CRSWNP | Control |
|
|
|
|
|
|
|||
| n | % | n | % | |||||||
| C | 56 | 90.32 | 133 | 91.1 | 0.031 | 0.316 | 0.992 | 3.891 | 0.912 | 2.528 |
| T | 6 | 9.68 | 13 | 8.9 | 0.031 | 0.684 | 1.009 | 1.886 | 1.096 | 3.023 |
| C/C | 25 | 80.65 | 61 | 83.56 | 0.129 | 0.333 | 0.965 | 4.051 | 0.820 | 2.419 |
| C/T | 6 | 19.35 | 11 | 15.07 | 0.292 | 0.287 | 1.284 | 5.303 | 1.353 | 4.048 |
Alleles C and T among patients with CRSwNP of the study group 1 and in the population group occurred with almost the same frequency. It was possible to note only a slight prevalence of the frequency of detection of genotype C among conventionally healthy individuals (χ2 = 0.03; р = 0.3; RR = 0.99; OR = 0.91; 95% CI: 3.891-2.52). There was also a slightly more significant, but still statistically insignificant, 1.08 times, excess of the T genotype frequency among patients with CRSwNP relative to the frequency of this genotype detection in the control sample (χ2 = 0.3; p = 0.6; RR = 1.0; OR = 1.09; 95% CI: 1.88-3.02). The frequency of detecting the C/C genotype of the C589T rs2243250 polymorphism in the IL4 gene is insignificant, less than 1.03 times, prevailed in the control group, relative to its values in group 1 (χ2 = 0.12; p = 0.33; RR = 0.96; OR = 0.82; 95% CI: 4.05-2.41). The frequency of occurrence of the C/T genotype differed among patients with CRSwNP exceeding 1.28 times, in the control group among conventionally healthy individuals, amounting to 19.35 and 15.07%, respectively (χ2 = 0.29; p = 0.28; RR = 1.28; OR = 1.35; 95% CI: 5.303-4.04) (Table 3).
Table 4 shows the results of a comparative analysis of the frequencies of detection of alleles and genotypes of the C589T rs2243250 polymorphism in the IL4 gene in group 2 and in the control group.
The frequency of occurrence of the C allele of the C589T rs2243250 polymorphism in the IL4 gene was statistically insignificant, 1.19 times higher among conventionally healthy individuals (χ2 = 9.3; p = 0.59; RR = 0.83; OR = 0.31; 95% CI: 1.675-0.65), and allele T indices were higher than 2.6 times in patients with CRS in group 2 (χ2 = 9.37; p = 0.40; RR = 1.19; OR = 3.18; 95% CI: 2.77-6.69). The C/C genotype in the population sample was detected 1.39 times more often than among patients with CRS of group 2, which was a statistically insignificant difference (χ2 = 7.6; p = 0.57; RR = 0.71; OR = 0.29; 95% CI: 1.79-0.69). The frequency of C/T genotype detection was statistically slightly significant, 1.9 times higher among patients with CRS than among conventionally healthy individuals (χ2 = 4.69; p = 0.3; RR = 2.15; OR = 2.71; 95% CI: 5.57-6.69). An analysis of the comparison of the occurrence of the T/T genotype showed a tendency to an increase in the frequency of its detection among patients with CRS, relative to the population sample, in which its values were 7.5% and 1.37%, respectively, 1.39 times more often among patients with CRS. 2 groups than among conditionally healthy individuals (χ2 = 1.37; p = 0.33; RR = 5.4; OR = 5.3; 95% CI: 18.57- 45.37) (Table 4).
| Table 4: Differences in the frequency of allelic and genotypic variants polymorphism C589T rs2243250 in the IL4 gene in patients with CRS and among conventionally healthy individuals. | ||||||||||
| Alleles and genotypes | Number of examined alleles and genotypes | Xi2 | p | RR | + 95%CI | OR | +95%CI | |||
| CRS | Control | |||||||||
| n | % | n | % | |||||||
| C | 61 | 76.25 | 133 | 91.1 | 9.372 | 0.594 | 0.837 | 1.675 | 0.314 | 0.659 |
| T | 19 | 23.75 | 13 | 8.9 | 9.372 | 0.406 | 1.195 | 2.773 | 3.187 | 6.694 |
| C/C | 24 | 60 | 61 | 83.56 | 7.697 | 0.571 | 0.718 | 1.792 | 0.295 | 0.699 |
| C/T | 13 | 32.5 | 11 | 15.07 | 4.694 | 0.303 | 2.157 | 5.572 | 2.714 | 6.697 |
| T/T | 3 | 7.5 | 1 | 1.37 | 2.844 | 0.339 | 5.475 | 18.579 | 5.838 | 45.379 |
Further, in the following table 5, the results of a comparative analysis of the frequencies of detection of alleles and genotypes of the polymorphic locus C589T rs2243250 in the IL4 gene among patients with CRSwNP and CRS of groups 1 and 2 are presented.
The incidence of the C allele was statistically slightly significantly higher among patients with CRSwNP by almost 1.18 times (χ2 = 4.7; p = 0.24; RR = 4.88; OR = 2.90; 95% CI: 4.88-7.57), and the T allele is insignificant was more often detected among patients with CRS (χ2 = 4.6; p = 0.76; RR = 0.84; OR = 0.34; 95% CI: 1.462-0.89). The frequency of C/C genotype detection was statistically insignificant, 1.34 times higher in patients with CRSwNP, compared with its values among patients with CRS (χ2 = 3.4; p = 0.27; RR = 1.34; OR = 2.77; 95% CI: 5.680-8.12). The values of the frequency of detection of the C/T genotype were slightly at a high level in patients with CRS and amounted to 19.35% and 32.5% (χ2 = 1.54; p = 0.48; RR = 0.59; OR = 0.49; 95% CI: 2.44 -1.49). Differences in the frequency of detection of the T/T genotype in groups 1 and 2 were statistically significant, there was a tendency to an increase in the value of this indicator among CRSwNP patients by 1.18 times, relative to its detectability among patients with CRS (χ2 = 4.7; p = 0.24; RR = 1.18; OR = 2.9; 95% CI: 4.88-7.57) (Table 5).
| Table 5: Differences in the frequency of allelic and genotypic variants polymorphism C589T rs2243250 in the IL4 gene among patients with CRSWNP and CRS. | ||||||||||
| Alleles and genotypes | Number of examined alleles and genotypes | Хi2 | p | RR | + 95%CI | OR | +95%CI | |||
| CRSWNP | CRS | |||||||||
| n | % | n | % | |||||||
| C | 56 | 90.32 | 61 | 76.25 | 4.769 | 0.240 | 1.185 | 4.885 | 2.907 | 7.576 |
| T | 6 | 9.68 | 19 | 23.75 | 4.769 | 0.760 | 0.844 | 1.462 | 0.344 | 0.896 |
| C/C | 25 | 80.65 | 24 | 60 | 3.481 | 0.273 | 1.344 | 5.680 | 2.778 | 8.126 |
| C/T | 6 | 19.35 | 13 | 32.5 | 1.540 | 0.481 | 0.596 | 2.442 | 0.498 | 1.498 |
| T/T | 56 | 90.32 | 61 | 76.25 | 4.769 | 0.240 | 1.185 | 4.885 | 2.907 | 7.576 |
Conclusion
Thus, we found that in patients with CRSwNP, the unfavorable allele C of the C589T rs2243250 polymorphism in the IL4 gene is more common than among healthy individuals and patients with CRS. There is a high frequency of occurrence of this allele with a predominance of the homozygous C/C variant (from 1.0 to 1.28 times). At the same time, the differences between patients with CRSwNP, CRS and the control sample were noted at the level of a trend, and the trend had a borderline level of statistical significance. These data allow us to conclude that the C allele and the C/C genotype of the C589T rs2243250 polymorphism in the IL4 gene have a predisposing effect on the development and clinical course of CRSWNP. Since this polymorphism is located in the promoter region of the gene and refers to functional polymorphisms. The presence of the C allele in patients with CRSwNP is accompanied by a decrease in the production of the IL-4 gene in the presence of the C/C genotype. The pattern of the inflammatory response gene is able to modify the implementation of the immune and inflammatory response of the facial skin towards an inappropriate hyperinflammatory response, leading to the progression and development of a more severe form of CRSwNP.
References
- Fokkens WJ, Lund VJ, Hopkins C, Hellings PW, Kern R, Reitsma S, Toppila-Salmi S, Bernal-Sprekelsen M, Mullol J, Alobid I, Terezinha Anselmo-Lima W, Bachert C, Baroody F, von Buchwald C, Cervin A, Cohen N, Constantinidis J, De Gabory L, Desrosiers M, Diamant Z, Douglas RG, Gevaert PH, Hafner A, Harvey RJ, Joos GF, Kalogjera L, Knill A, Kocks JH, Landis BN, Limpens J, Lebeer S, Lourenco O, Meco C, Matricardi PM, O’Mahony L, Philpott CM, Ryan D, Schlosser R, Senior B, Smith TL, Teeling T, Tomazic PV, Wang DY, Wang D, Zhang L, Agius AM, Ahlstrom-Emanuelsson C, Alabri R, Albu S, Alhabash S, Aleksic A, Aloulah M, Al-Qudah M, Alsaleh S, Baban MA, Baudoin T, Balvers T, Battaglia P, Bedoya JD, Beule A, Bofares KM, Braverman I, Brozek-Madry E, Richard B, Callejas C, Carrie S, Caulley L, Chussi D, de Corso E, Coste A, El Hadi U, Elfarouk A, Eloy PH, Farrokhi S, Felisati G, Ferrari MD, Fishchuk R, Grayson W, Goncalves PM, Grdinic B, Grgic V, Hamizan AW, Heinichen JV, Husain S, Ping TI, Ivaska J, Jakimovska F, Jovancevic L, Kakande E, Kamel R, Karpischenko S, Kariyawasam HH, Kawauchi H, Kjeldsen A, Klimek L, Krzeski A, Kopacheva Barsova G, Kim SW, Lal D, Letort JJ, Lopatin A, Mahdjoubi A, Mesbahi A, Netkovski J, Nyenbue Tshipukane D, Obando-Valverde A, Okano M, Onerci M, Ong YK, Orlandi R, Otori N, Ouennoughy K, Ozkan M, Peric A, Plzak J, Prokopakis E, Prepageran N, Psaltis A, Pugin B, Raftopulos M, Rombaux P, Riechelmann H, Sahtout S, Sarafoleanu CC, Searyoh K, Rhee CS, Shi J, Shkoukani M, Shukuryan AK, Sicak M, Smyth D, Sindvongs K, Soklic Kosak T, Stjarne P, Sutikno B, Steinsvag S, Tantilipikorn P, Thanaviratananich S, Tran T, Urbancic J, Valiulius A, Vasquez de Aparicio C, Vicheva D, Virkkula PM, Vicente G, Voegels R, Wagenmann MM, Wardani RS, Welge-Lussen A, Witterick I, Wright E, Zabolotniy D, Zsolt B, Zwetsloot CP. European Position Paper on Rhinosinusitis and Nasal Polyps 2020. Rhinology. 2020 Feb 20;58(Suppl S29):1-464. doi: 10.4193/Rhin20.600. PMID: 32077450.
- Levchenko AS, et al. Genetic aspects of chronic rhinosinusitis. Russian Journal of Genetics. 2018;910-918.
- Hamilos DL. Chronic Rhinosinusitis in Patients with Cystic Fibrosis. J Allergy Clin Immunol Pract. 2016 Jul-Aug;4(4):605-12. doi: 10.1016/j.jaip.2016.04.013. PMID: 27393775.
- Chakrabarti A, Kaur H. Allergic Aspergillus Rhinosinusitis. J Fungi (Basel). 2016 Dec 8;2(4):32. doi: 10.3390/jof2040032. PMID: 29376948; PMCID: PMC5715928.
- Miłoński J, Zielińska-Bliźniewska H, Przybyłowska K, Pietkiewicz P, Korzycka-Zaborowska B, Majsterek I, Olszewski J. Significance of CYCLOOXYGENASE-2(COX-2), PERIOSTIN (POSTN) and INTERLEUKIN-4(IL-4) gene expression in the pathogenesis of chronic rhinosinusitis with nasal polyps. Eur Arch Otorhinolaryngol. 2015 Dec;272(12):3715-20. doi: 10.1007/s00405-014-3481-9. Epub 2015 Jan 9. PMID: 25573836; PMCID: PMC4633441.
- Schleimer RP. Immunopathogenesis of Chronic Rhinosinusitis and Nasal Polyposis. Annu Rev Pathol. 2017 Jan 24;12:331-357. doi: 10.1146/annurev-pathol-052016-100401. Epub 2016 Dec 5. PMID: 27959637; PMCID: PMC5514544.
- Mohamad S, Hamid SSA, Azlina A, Md Shukri N. Association of IL-1 gene polymorphisms with chronic rhinosinusitis with and without nasal polyp. Asia Pac Allergy. 2019 Jul 8;9(3):e22. doi: 10.5415/apallergy.2019.9.e22. PMID: 31384577; PMCID: PMC6676066.
- Zhang Q, Wang X, Cheng X, Wu X, Feng Y, Xu H, Zhu H, Yi H, Zhang W, Li X, Ye H. Multiple genetic variations of chronic rhinosinusitis with nasal polyps are associated with respiratory parameters in men with obstructive sleep apnea. Sleep Breath. 2021 Mar 26. doi: 10.1007/s11325-021-02356-6. Epub ahead of print. PMID: 33770380.
- Kondratyeva E, Efremova A, Melyanovskaya Y, Petrova N, Satsuk N, Bulatenko N, Bukharova T, Zodbinova A, Sherman V, Kashirskaya N, Zinchenko R, Kutsev S, Goldshtein D. Clinical and genetic characterization of patients with cystic fibrosis and functional assessment of the chloride channel with the pathogenic variant c.831G>A (p.Trp277*), described for the first time. Gene. 2020 Nov 30;761:145023. doi: 10.1016/j.gene.2020.145023. Epub 2020 Aug 3. PMID: 32758581.
- Xiong G, Xie X, Wang Q, Zhang Y, Ge Y, Lin W, Li M. Immune cell infiltration and related core genes expression characteristics in eosinophilic and non-eosinophilic chronic rhinosinusitis with nasal polyps. Exp Ther Med. 2020 Dec;20(6):180. doi: 10.3892/etm.2020.9310. Epub 2020 Oct 12. PMID: 33101470; PMCID: PMC7579783.
- Lam K, Schleimer R, Kern RC. The Etiology and Pathogenesis of Chronic Rhinosinusitis: a Review of Current Hypotheses. Curr Allergy Asthma Rep. 2015 Jul;15(7):41. doi: 10.1007/s11882-015-0540-2. PMID: 26143392; PMCID: PMC4874491.
- Pietruszewska W, Fendler W, Podwysocka M, Białas AJ, Kuna P, Kupryś-Lipińska I, Borowiec M. Expression of Transcript Variants of PTGS1 and PTGS2 Genes among Patients with Chronic Rhinosinusitis with Nasal Polyps. Diagnostics (Basel). 2021 Jan 16;11(1):135. doi: 10.3390/diagnostics11010135. PMID: 33467191; PMCID: PMC7830232.
- Milonski J, Zielinska-Blizniewska H, Majsterek I, Przybyłowska-Sygut K, Sitarek P, Korzycka-Zaborowska B, Olszewski J. Expression of POSTN, IL-4, and IL-13 in Chronic Rhinosinusitis with Nasal Polyps. DNA Cell Biol. 2015 May;34(5):342-9. doi: 10.1089/dna.2014.2712. Epub 2015 Feb 12. PMID: 25674935.
- Khalmuratova R, Lee M, Park JW, Shin HW. Evaluation of Neo-Osteogenesis in Eosinophilic Chronic Rhinosinusitis Using a Nasal Polyp Murine Model. Allergy Asthma Immunol Res. 2020 Mar;12(2):306-321. doi: 10.4168/aair.2020.12.2.306. PMID: 32009324; PMCID: PMC6997277.
- Nikakhlagh S, et al. Evaluation of C-590T Promoter of IL-4 Gene Polymorphisms in Patients with Sinonasal Polyposis //International journal of pharmaceutical research and allied sciences. 2016;39-43.
- Koennecke M, Benecke F, Masche A, Linke R, Bruchhage KL, Pries R, Klimek L, Wollenberg B. Increased phosphorylation of eNOS in nasal polyps of chronic rhinosinusitis patients can be diminished by 1,8-cineol. Nitric Oxide. 2018 Aug 1;78:89-94. doi: 10.1016/j.niox.2018.06.002. Epub 2018 Jun 6. PMID: 29885366.
- Akyigit A, Keles E, Etem EO, Ozercan I, Akyol H, Sakallioglu O, Karlidag T, Polat C, Kaygusuz I, Yalcin S. Genetic polymorphism of antioxidant enzymes in eosinophilic and non-eosinophilic nasal polyposis. Eur Arch Otorhinolaryngol. 2017 Jan;274(1):267-273. doi: 10.1007/s00405-016-4259-z. Epub 2016 Aug 11. PMID: 27515707
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.